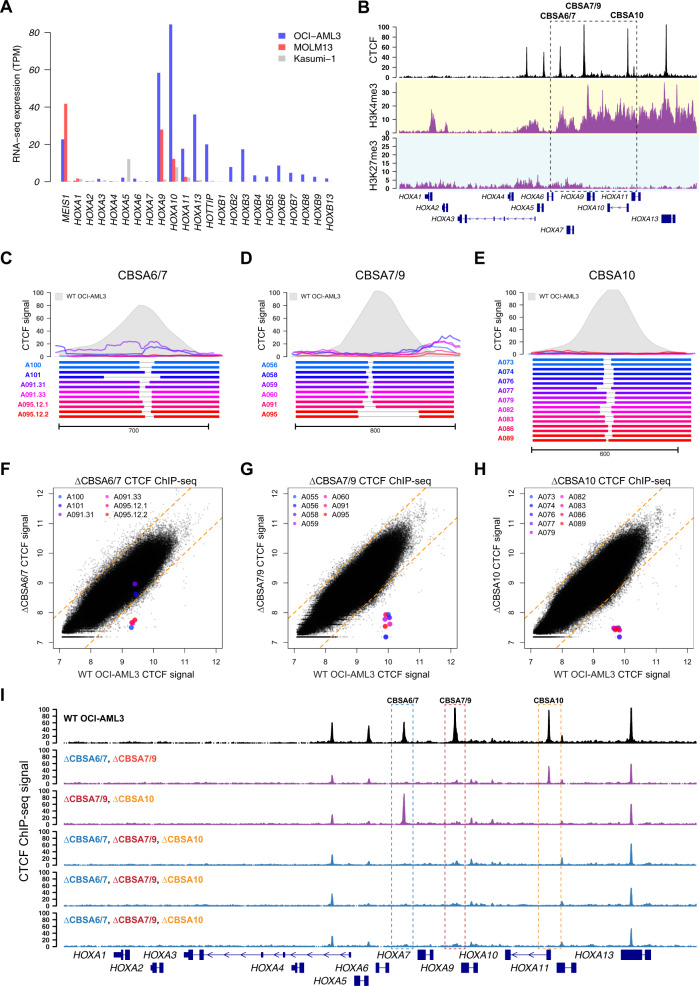
Fig. 3. Targeted deletions eliminate CTCF binding in the NPM1-mutant OCI-AML3 cell line.
a RNA-seq expression of HOXA and HOXB genes in OCI-AML3 cells, which display the canonical mutant NPM1-associated HOXA/HOXB expression phenotype. Also shown are the MLL-rearranged MOLM13 cell line that expresses only HOXA genes, and the RUNX1-RUNX1T1-containing Kasumi-1 cell line that has low HOXA and HOXB gene expression. b ChIP-seq data from OCI-AML3 cells for CTCF (black), H3K4me3 (highlighted in yellow), and H3K27me3 (highlighted in blue), which show conserved CTCF binding sites and distinct regions of active (H3K4me3) and repressed (H3K27me3) chromatin. c–e Targeted deletions that disrupt CTCF binding in OCI-AML3 cells at sites CBSA6/7 (in c), CBSA7/9 (in d), and CBSA10 (in e). Bottom panels show allele pairs from homozygous or compound heterozygous deletion mutants at each site; top panels show CTCF ChIP-seq signal from these mutant cell lines (multi-colored lines) compared with wild-type OCI-AML3 cells (in gray). f–h CTCF ChIP-seq signal (log2 normalized read counts) for all CTCF peaks from deletion mutants (Y axis) vs. wild-type OCI-AML3 cells, showing dramatically reduced CTCF ChIP-seq signal in deletion mutants at all three sites, with the exception of clones A101 and A091.31, which only partially eliminates CTCF signal at site CBSA6/7. i CTCF ChIP-seq tracks from double (in purple) and triple mutants (in blue), generated via sequential-targeted deletion experiments. CTCF ChIP-seq from wild-type OCI-AML3 cells is shown in black at the top for reference.