Figure 6.
A conserved cation-sensitive network necessary for DISP1 and PTCH1 function
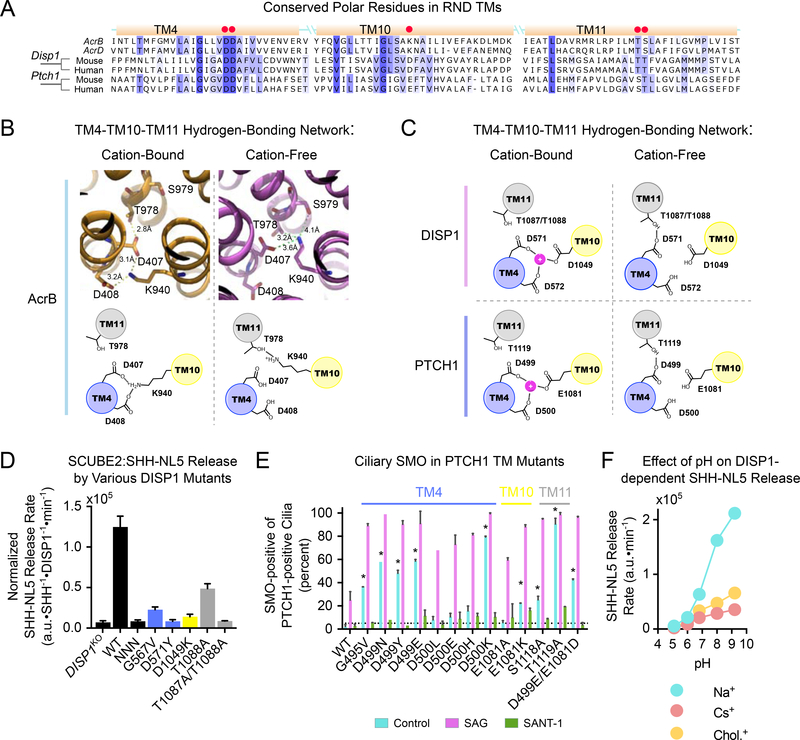
(A) Sequence alignment of DISP1, PTCH1, and E. coli AcrB and AcrD. TM helices are shown as yellow boxes, with red dots indicating polar residues necessary for DISP1, PTCH1 and AcrB function.
(B) Proton-dependent conformational changes in AcrB (PDB ID: 2gif). TM4-resident D407 and D408 residues alternate between being deprotonated and bound to the TM10-resident K940 (left), and being protonated and with K940 bound to T978/S979 in TM11 (right).
(C) Proposed model of cation-dependent conformational switching in DISP1 and PTCH1. TM10 hosts an aspartate in DISP1 and glutamate in PTCH1, instead of a lysine. This suggests that the deprotonated acidic residues in TM4 and TM10 form a cation-binding site. In the absence of cation, the first TM4 aspartate binds to the S/T cluster in TM11, to effect conformational change. Residue numbers are for mouse DISP1 and PTCH1.
(D) TM-resident polar residues in DISP1 are necessary for SHH release. DISP1KO HEK293T cells stably expressing SHH-NL5 were stably rescued with DISP1 mutants, and SCUBE2-dependent SHH-NL5 release was measured. Protein synthesis was inhibited with CHX (100μg/mL). Data show estimate of release rate normalized to DISP1 and SHH-NL5 levels in single biological replicate; error bars represent combined standard error and standard deviation around the mean.
(E) TM-resident polar residues in PTCH1 are necessary for SMO repression. PTCH1 mutants were stably expressed in Ptch1−/− MEFs, and their ability to reduce ciliary levels of endogenous SMO was assayed by immunofluorescence. SAG (500nM) and SANT-1 (1μM) served as positive and negative control, respectively. Error bars represent standard deviation around the mean of two biological replicates. At least 100 cilia were assayed per replicate. * p<0.01, unpaired, two-tailed t-test.
(F) As in (D), but HEK293T cells stably expressing SHH-NL5 were incubated with SCUBE2 (lμM), in modified Tyrode’s media with high Na+, choline+ or Cs+, buffered to different pH values. SHH-NL5 release is potentiated by increasing pH, but only if the Na+ gradient is present. Error bars are obscured, but indicate standard error of the estimate in single replicate.
See also Figure S5 for evidence of an alternating hydrogen-bonding network in PTCH1, similar to AcrB. See also Figure S6 for characterization of cells used for DISP1 and PTCH1 mutant analysis.