Figure 3: The trophectoderm is patterned by a spatial and temporal ERK activity gradient.
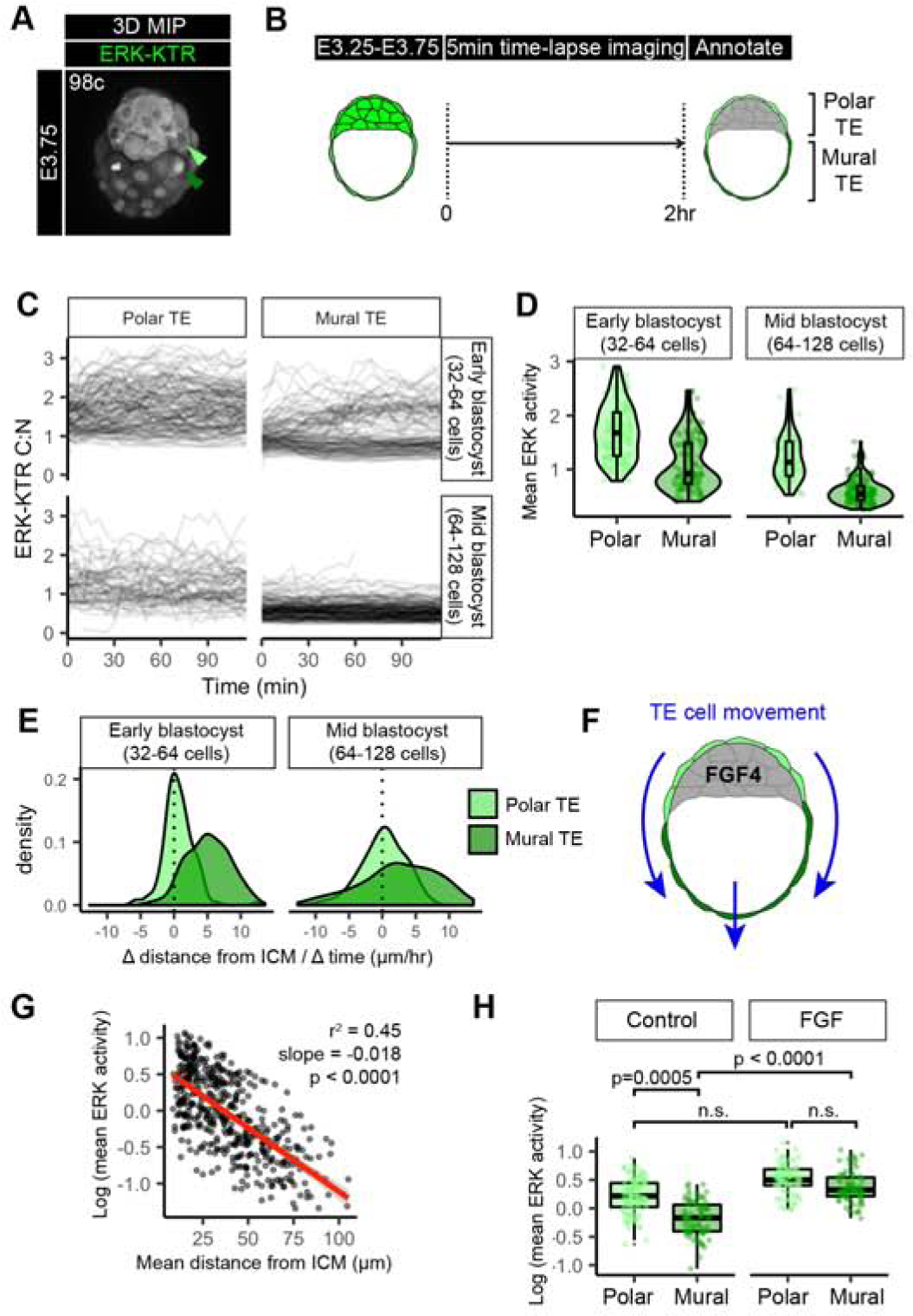
(A) Confocal image of hemizygous HprtERK-KTR and homozygous R26NLS-mKate2 embryos during preimplantation development. Embryonic day (E) and cell number are indicated for staging. Reproduced from panel within Figure 1D. (B) Schematic of time-lapse confocal imaging where polar versus mural TE identity was annotated by position at the end of the movie. (C) Traces of single cell ERK-KTR C:N values over the course of the 2 hour time-lapse imaging. Single-cell traces are shown in black, for polar TE, and mural TE. Stage of embryo at the beginning of the movie is indicated, early blastocyst (32–64 cell) stage and mid-blastocyst (64–128 cell) stage. (D) Violin and boxplots showing mean ERK activities in individual TE cells over the 2 hour time-lapse imaging. Mean ERK-KTR C:N values per cell are shown for every cell as individual points. Polar TE: light green, Mural TE: dark green. Cell stages at beginning of time-lapse are indicated. early-blastocyst stage: n=14, mid-blastocyst stage: n=10 embryos. (E) Density plots showing the change (Δ) in distance from the nearest ICM cell over the duration of the time-lapse for individual Polar TE and Mural TE cells. (F) Schematic of TE movement along the embryonic – abembryonic axis away from the inner cell mass (ICM, grey) derived FGF4 signals. (G) Scatterplot comparing the mean distance over the lifetime of that cell to its nearest ICM neighbor in each frame, with the mean ERK activity of that cell. Linear regression line of best fit shown in red. Slope, adjusted r2 goodness of fit, and p value are indicated. (H) Boxplot showing mean ERK activities in individual ICM cells over the 2 hour time-lapse imaging. Mean ERK-KTR C:N values per cell are shown for every cell as individual points. Color coding for TE sub-types as indicated before. Control: n=10, FGF: n=8 embryos (as in Figure 2). P values for statistical significance of pairwise comparisons are shown (Wilcoxon test), horizontal lines indicate groups compared. Not significant (n.s.) pairwise comparisons indicated where p > 0.05.
