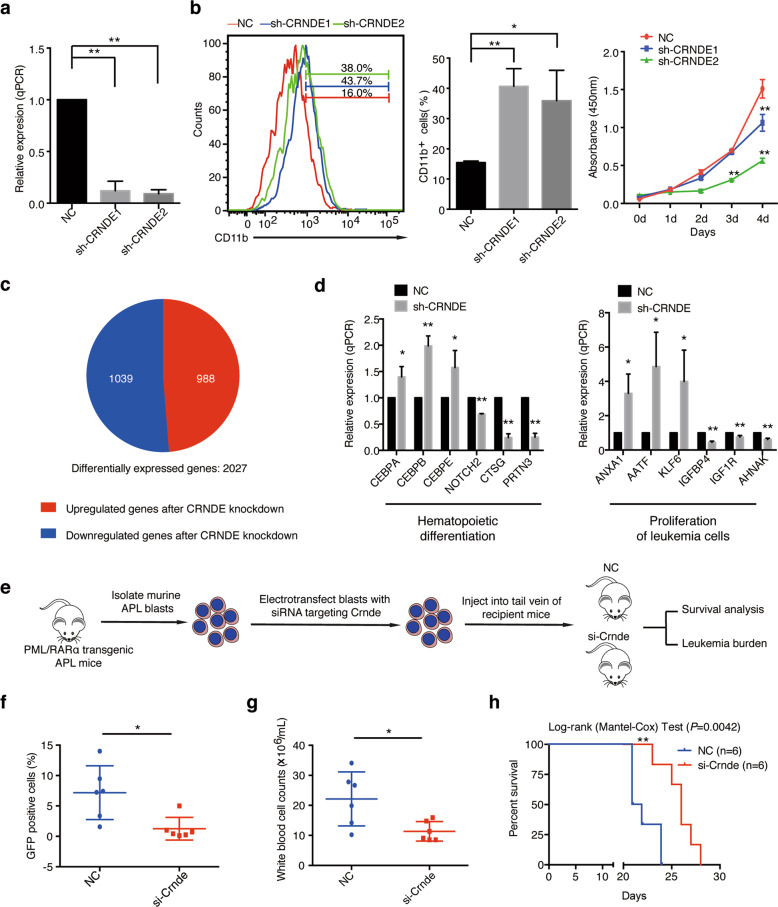
Fig. 3. CRNDE knockdown induces differentiation and inhibits proliferation in vitro and suppresses APL leukemogenesis in vivo.
a The knockdown efficiency of CRNDE was detected in NB4 cells by qRT-PCR after the transfection of CRNDE shRNAs for 72 h. The expression level of GAPDH was used for normalization of qRT-PCR data, and bars represent the mean (s.d.) of three independent experiments. **P < 0.01. b Functional analysis was performed after CRNDE knockdown in NB4 cells. Differentiation of NB4 cells was determined by flow cytometric analysis of CD11b expression after transfection with indicated shRNAs, and flow cytometric data was displayed as histogram (left panel) and bar graph (middle panel). The bar graph shows the average percentage of CD11b-positive cells derived from three independent experiments, and data are represented as the mean ± s.d. The proliferation of NB4 cells was detected by CCK8 cell viability assays after transfection with shRNAs at the indicated time (right panel). *P < 0.05, **P < 0.01. c Pie chart presents the proportion of differentially expressed genes (fold change > 1.5) based on RNA-seq data after the transfection of CRNDE shRNA for 72 h. d qRT-PCR assays were performed to validate differentially expressed genes after CRNDE knockdown in NB4 cells. *P < 0.05, **P < 0.01. e The schematic plot represents the procedure performed in vivo. The leukemic blasts from PML/RARα transgenic APL mice were electrotransfected with siRNA targeting Crnde, and then injected into tail vein of the recipient FVB/NJ mice. f–g The leukemia burden was detected in the peripheral blood of PML/RARα transgenic APL mice with or without Crnde knockdown. (f) shows the total white blood cell counts, and (g) represents the percentage of GFP-positive APL cells. *P < 0.05. h Kaplan–Meier curve shows the survival rates of mice receiving APL blasts transfected with irrelevant control (NC) or siRNA targeting Crnde (n = 6 for each model). P-value was analyzed by a log-rank (Mantel–Cox) test. **P < 0.01.