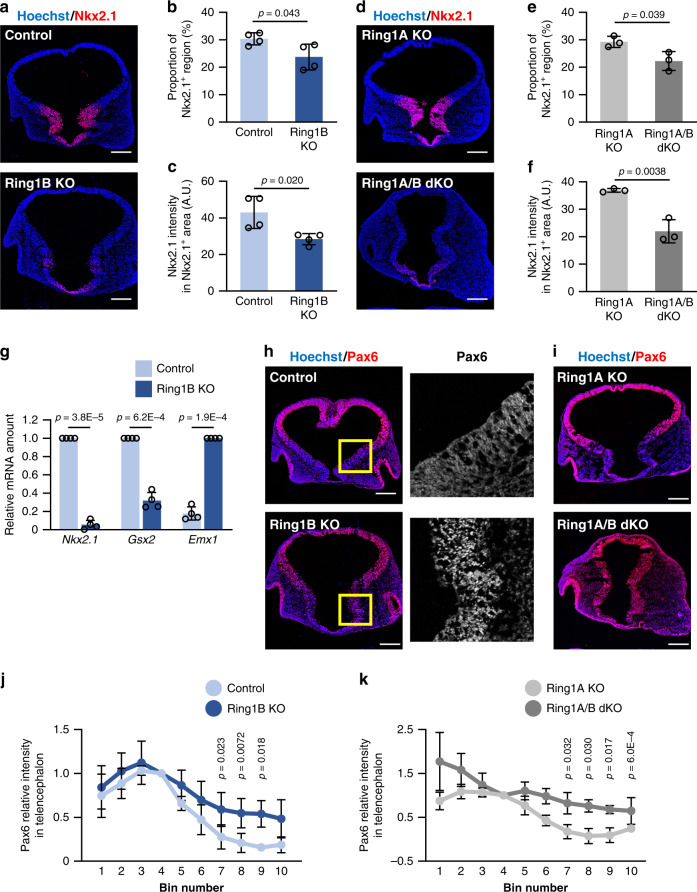
Fig. 2. Deletion of Ring1 confers a dorsal-like expression pattern of region-specific TFs in NPCs of the ventral telencephalon.
a, d, h, i Coronal sections of the brain of control and Ring1B KO mice (a, h) or of Ring1A KO and Ring1A/B dKO mice (d, i) at E10 were subjected to immunohistofluorescence staining with antibodies to Nkx2.1 (a, d) or to Pax6 (h, i). Nuclei were counterstained with Hoechst 33342. The regions indicated with yellow outlined squares (300 by 300 μm) in the left panels of (h) are shown at higher magnification in the right panels. Scale bars, 200 μm. b, c, e, f Quantification of immunostaining for Nkx2.1 in images similar to those in (a) and (d). The Nkx2.1+ perimeter length as a proportion of total perimeter length for the telencephalic wall (b, e) as well as the average Nkx2.1 immunostaining intensity (in arbitrary units, A.U.) within the Nkx2.1+ region (pixels) for each section (c, f) were determined. Data are means ± s.d., n = 4 (b, c) and n = 3 (e, f) embryos of each genotype, two-tailed Student’s unpaired t test. g Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR) analysis of relative Nkx2.1, Gsx2, or Emx1 mRNA abundance (normalized by the amount of Actb mRNA) in CD133+ NPCs isolated from the GE of control or Ring1B KO mice at E11. Data are means ± s.d., averaged values for n = 4 litters (the number and individual values of each littermate are provided in Source Data File), two-tailed Student’s paired t test. j, k Quantification of immunostaining intensity of Pax6 for images similar to those in (h) and (I), respectively. The telencephalic wall was divided into 10 bins, from 1 (dorsal) to 10 (ventral), and the average immunostaining intensity of Pax6 was determined in each bin and normalized by the average value for bin 4. Data are means ± s.d., n = 4 embryos from each genotype, two-tailed Student’s paired t test between values in each bin. Source data are provided as a Source Data file.