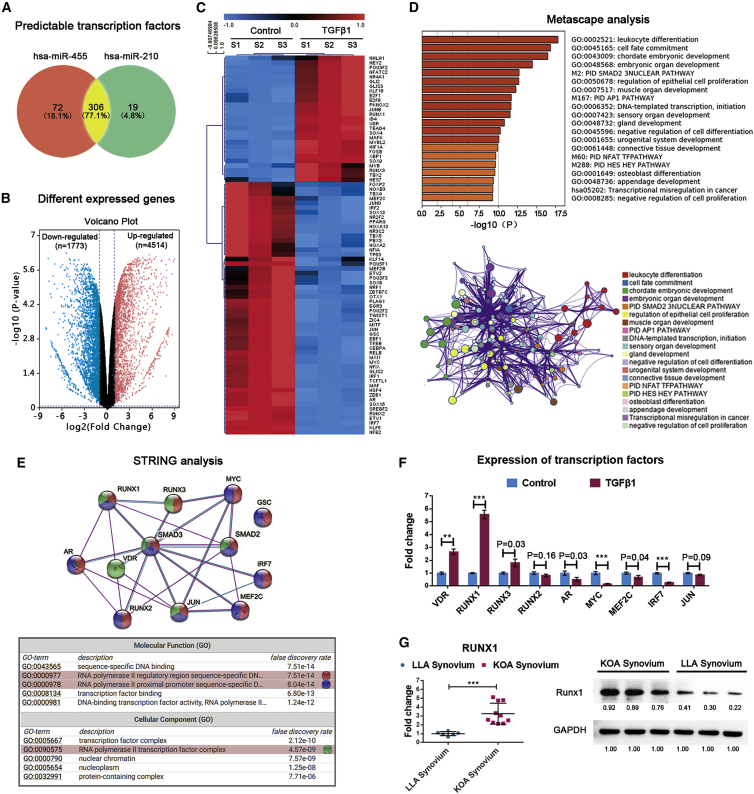
Figure 5.
Prediction of TFs for miR-455 and miR-210
(A) Venn diagrams showing the numbers of predicted TFs for miR-455 and miR-210. Transcriptome sequencing was conducted on synoviocytes from three KOA patients. Volcano plots of DEGs in synoviocytes treated with or without TGF-β1 (10 ng/mL). (B) Red indicates significantly upregulated genes, blue indicates significantly downregulated genes, and gray indicates genes with no differential expression based on the criteria of p < 0.05 and fold change > 2.0. (C) Heatmap of the predicted TFs that were significantly changed in TGF-β1-activated synoviocytes. Pathways significantly associated with these potential TFs are shown. (D) The enriched terms are depicted in a heatmap and an interaction network according to their p values. (E) A protein-protein interaction (PPI) network of the TFs involved in the Smad2/3 pathway was constructed, and the corresponding GO terms were mapped. (F) The expression patterns of Smad2/3 pathway-related TFs in synoviocytes treated with TGF-β1 (10 ng/mL) as determined by qRT-PCR. Synoviocytes from KOA patients were used for the above cell experiments. (G) Left: Runx1 expression changes in synovial tissue from KOA patients (n = 10) and LLA patients (n = 6) as detected by qRT-PCR. Right: the Runx1 levels in three random samples from each group were validated by western blot analysis. The qRT-PCR data are presented as the mean and SD, and representative images of western blots are shown. GAPDH was detected as an endogenous control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.