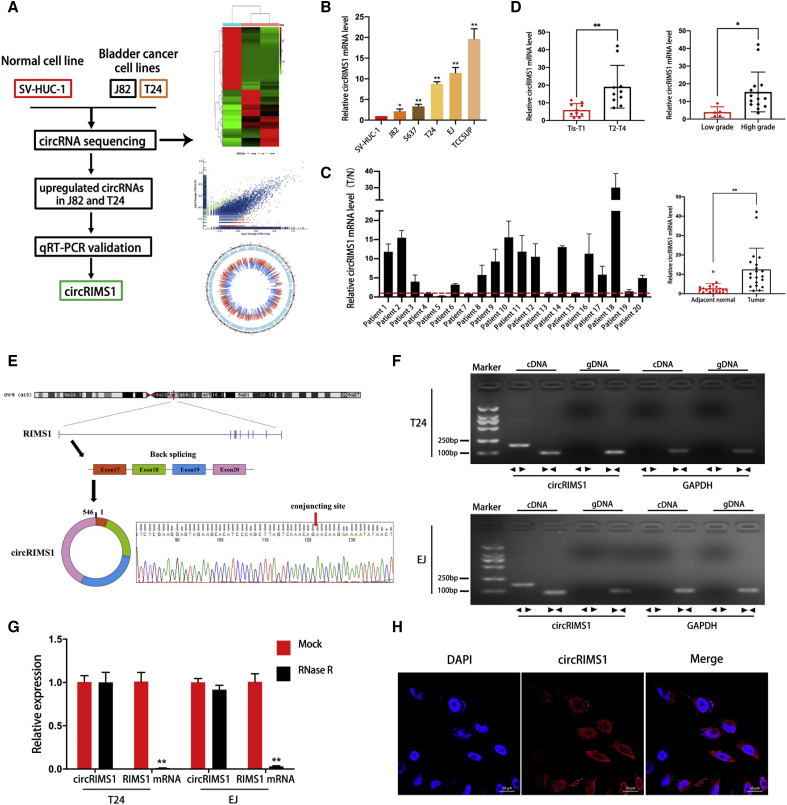
Figure 1.
circRNA Expression Profiles and Validation of circRIMS1 in Bladder Cancer Tissues and Cells
(A) A flow diagram of circRNA sequencing in SV-HUC-1, J82, and T24 cell lines. (B) Levels of circRIMS1 in SV-HUC-1 cells and bladder cancer cells (J82, 5637, T24, EJ, and TCCSUP) were determined by qRT-PCR. circRIMS1 was upregulated in bladder cancer cell lines. (C) Analysis of circRIMS1 expression fold change in tumor tissues and adjacent normal tissues of bladder cancer patients (n = 20). (D) Expression of circRIMS1 in our bladder cancer patients during different pathological stages and histological grades. (E) Schematic illustration indicating the circularization of exons 17, 18, 19, and 20 of RIMS1, forming circRIMS1. RT-PCR was performed to verify the existence of circRIMS1. Sanger sequencing further proved the head-to-tail splice site in circRIMS1, and the specific junction is indicated by the red arrow. (F) circRIMS1 was detected in T24 and EJ cell lines by RT-PCR. Divergent primers could only amplify circRIMS1 from cDNA. GAPDH was used as a negative control. (G) circRIMS1 and linear RIMS1 mRNA levels in T24 and EJ cells were determined by qRT-PCR, with or without RNase R. (H) RNA FISH for circRIMS1 was detected in EJ cells. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue), and circRIMS1 appeared red. Results indicated that circRIMS1 was primarily localized in the cytoplasm. Scale bars, 20 μm. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 versus control group.