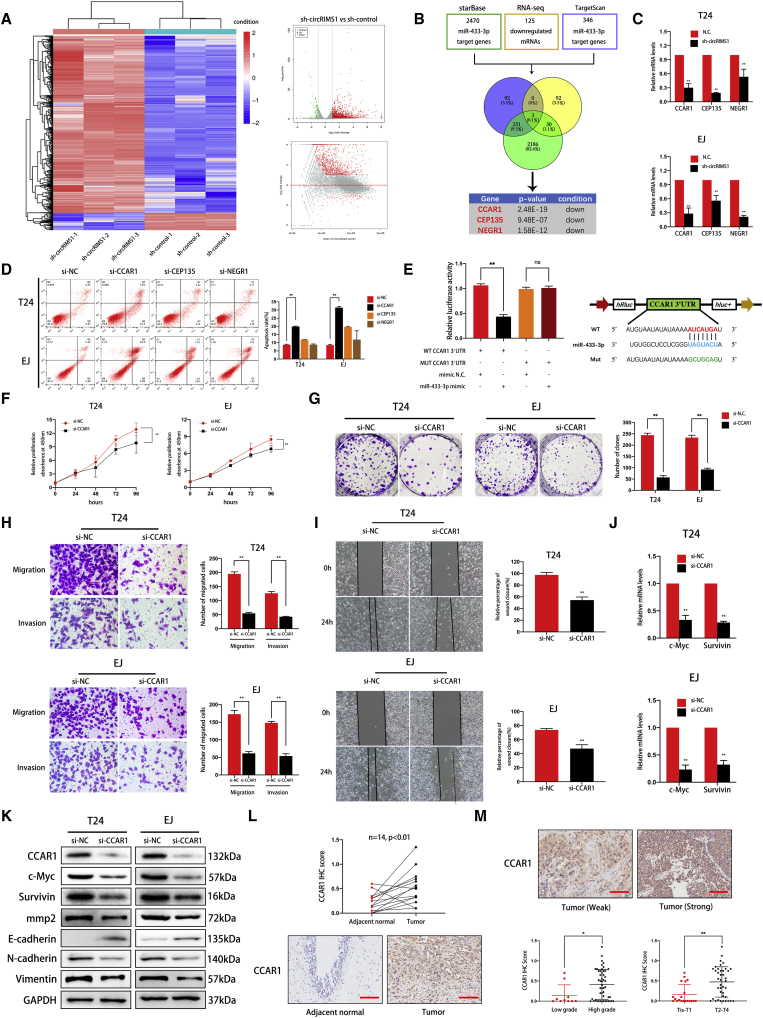
Figure 6.
CCAR1 Is Directly Targeted by miR-433-3p and Serves as a Tumor Promoter in Bladder Cancer
(A) Significantly differentially expressed mRNAs are presented as a clustered heatmap in T24 cells (negative control versus sh-circRIMS1). Red dots represent upregulated genes, and green dots indicate downregulated genes in the volcano plot. (B) Overlap of downregulated mRNAs identified by RNA sequencing and target genes of miR-433-3p predicted from TargetScan and StarBase databases. (C) Expression levels of CCAR1, CEP135, and NEGR1 in control or circRIMS1-knockdown cells were further verified. (D) The biological effect of CCAR1 was determined by siRNA transfections and Annexin V-FITC/PI staining. (E) Luciferase reporter assay confirmed the interaction between miR-433-3p and the CCAR1 3′ UTR. miR-433-3p mimics (or negative control mimic) were cotransfected with wild-type or mutated CCAR1 3′ UTR luciferase reporter vector into HEK293 cells. (F and G) Cell viability and colony-forming ability in control or CCAR1-deficient cells evaluated by CCK-8 assay (F) and colony-formation assay (G), respectively. (H and I) To confirm the role of CCAR1 in cell migration and invasion abilities, Transwell migration and invasion assays (H) and wound-healing assay (I) were used to detect cell migration ability. (J) mRNA expression levels of two Wnt signaling target genes (c-Myc and survivin) were downregulated by CCAR1 depletion, as determined by qRT-PCR. (K) Expression of CCAR1, c-Myc, survivin, MMP2, and EMT markers at the protein level by western blotting in cells with various treatments. (L and M) Immunohistochemistry (IHC) detection of CCAR1 in tissue chip (n = 60). Representative images are shown, and IHC score was calculated from immunostaining intensity and positive staining rate. CCAR1 was abundantly expressed in bladder cancer tissue (L) and was associated with pathological stage and histological grade (M). Scale bars, 100 μm. ∗p < 0.05, ∗∗p < 0.01 versus control group.