Abstract
Research suggested accumulation of tau proteins might lead to the degeneration of functional networks. Studies investigating the impact of genetic risk for Alzheimer’s disease (AD) on early brain connections might shed light on mechanisms leading to AD development later in life. Here, we aim to investigate whether the polygenic risk score for Alzheimer’s disease (AD-PRS) influences the connectivity among regions susceptible to tau pathology during childhood and adolescence. Participants were youth, aged 6–14 years, and recruited in Porto Alegre (discovery sample, n = 332) and São Paulo (replication sample, n = 304), Brazil. Subjects underwent genotyping and 6-min resting state funcional magnetic resonance imaging. Connections between the local maxima of tau pathology networks were used as dependent variables. The AD-PRS was associated with the connectivity between the right precuneus and the right superior temporal gyrus (discovery sample: β = 0.180, padjusted = 0.036; replication sample: β = 0.202, p = 0.031). This connectivity was also associated with inhibitory control (β = 0.157, padjusted = 0.035) and moderated the association between the AD-PRS and both immediate and delayed recall. These findings suggest the AD-PRS may affect brain connectivity in youth, which might impact memory performance and inhibitory control in early life.
Keywords: Alzheimer’s disease, Polygenic risk score, Tau protein, Functional connectivity
1. Introduction
The accumulation of neurofibrillary tangles of hyper phosphorylated tau is one of the earliest and most important features of Alzheimer’s disease (AD), leading to neuronal damage and impaired cognition (Nordberg, 2015). It has been hypothesized that this process progresses along functional brain networks for decades before disease onset (Palop et al., 2006; Seeley et al., 2009), causing variations in their activity and leading to clinical manifestations later in life. Understanding how genetic risk for AD affects brain connectivity in youth might shed light on mechanisms leading to AD development later in life.
Previous evidence has shown brain regions exhibiting excessive tau accumulation coalesce into 10 tau pathology networks, which have been associated with clinical outcomes, such as disease stage and global cognition dysfunction, and with segments of several well-established functional networks, such as default mode network and language network (Hoenig et al., 2018). Examining the connections among regions susceptible to accumulate the tau protein is particularly important given that AD seems to affect several higher-order cognitive networks (Badhwar et al., 2017) but is not specific to any large-scale functional brain network as a whole (Hansson et al., 2017). This might be partially explained by regional differences in the vulnerability to tau accumulation (Hansson et al., 2017). These abnormalities in brain connections have been found not only within functional networks but also among them, leading to the hypothesis of a disconnection syndrome (Elman et al., 2016; Liao et al., 2018; Wang et al., 2015).
Abnormalities associated with AD appear to manifest decades before disease’s diagnosis (Braak and Del Tredici, 2011; Su et al., 2017). Postmortem studies found pre-tangle tau alterations (stages a-c) in 25% of children aged up to 10 years and almost 70% of adolescents aged 10 to 20 in a few susceptible brain regions, whereas β amyloid plaques only appeared after ages 30–40 years (Braak et al., 2011). Furthermore, previous studies found that the presence of the apolipoprotein E (APOE) ε4 allele and family history of AD relate to impairments in functional brain networks in asymptomatic adults (Su et al., 2017; Wang et al., 2012). Nevertheless, to our knowledge, no previous study has investigated the impact of genetic susceptibility to AD in functional brain connectivity during childhood and adolescence.
Our study aimed to investigate the implications of the polygenic risk score for Alzheimer’s disease (AD-PRS) on the connectivity patterns among tau pathology networks’ nodes in 2 samples of children and adolescents. We also aimed to investigate the association of these connections with the APOE gene and with nondeclarative memory performance and executive function.
2. Materials and methods
2.1. Participants
Our samples were composed of children and adolescents from the Brazilian High-Risk Study for Psychiatric Disorders (Salum et al., 2015). Subjects were recruited in Porto Alegre (discovery sample) and São Paulo (replication sample), Brazil. Eligibility criteria were the following: (1) being 6–12 years old at enrollment and (2) being registered in school by a biological parent who was a primary carer and could provide information about their children’s behavior. After a screening phase (n = 9937), a high-risk subgroup for psychiatric disorders (n = 2371) and a random-selection subgroup (n = 1500) were selected. In the high-risk subgroup, 14% did not fulfill inclusion criteria and 24% refused to participate or lost contact; in the random-selection group, these numbers were 12% and 27%, respectively. Finally, 958 (64%) and 1554 (66%) subjects completed household evaluation in the high-risk subgroup and the random-selection subgroup, respectively. A subsample of 636 subjects (332 in Porto Alegre and 304 in São Paulo) underwent genotyping and funcional magnetic resonance imaging acquisition in 2010. Participants and parents provided written or verbal consent. This study was approved by the Ethics Committee of the University of São Paulo and of the Hospital de Clinicas de Porto Alegre. Further information about participant selection can be found elsewhere (Salum et al., 2015).
2.2. Genotyping and polygenic risk score
After blood collection in EDTA tubes, we isolated genomic DNA using GentraPuregene Kit (Qiagen). We performed genotyping using the HumanOmniExpress V1 (Illumina), excluding single-nucleotide polymorphisms (SNPs) with a minor allele frequency < 1%, locus missingness > 10%, or Hardy-Weinberg equilibrium significance < 0.000001 and subjects with genotype missingness > 10% and estimation of identity by descent greater than 0.12. Genotype imputation was performed in https://imputationserver.sph.umich.edu, using the 1000G Phase 1 v3 and the Pre-phasing algorithm SHAPEIT2.
We calculated the AD-PRS using the PRSice software (Euesden et al., 2015), based on the summary statistics of the International Genomics of Alzheimer’s Project (Lambert et al., 2013; available at http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php). P-value-informed clumping was performed retaining the SNP with the smallest p-value within a 250-kb window and excluding SNPs in linkage disequilibrium (r2 > 0.1). We selected a priori a p-value threshold of <0.01 (nSNPs = 5116), in keeping with previous studies (Axelrud et al., 2018; Mormino et al., 2016).
To investigate APOE polymorphisms, we extracted a total of 19 SNPs within the APOE gene including a window of 500 bp upstream and downstream (HG19 chr19: 45, 408, 539–45, 413, 150). The APOE gene was not included in the AD-PRS.
2.3. Neuroimaging
Images were acquired using 1.5-T functional magnetic resonance image systems (GE Signa HD in the discovery sample and GE Signa HDX in the replication sample), with the following parameters: TR = 2000 ms, TE = 30 ms, slice thickness = 4 mm, gap = 0.5 mm, flip angle = 80°, matrix size = 80 × 80, NEX = 1, slices = 26. We used a resting-state protocol, in which subjects looked at a fixation point, with a total acquisition time of 6 minutes. Data were processed in AFNI (version 2011_12_21_1014) and FSL (version 5.0) packages.
The preprocessing consisted of several steps, as previously described (Sato et al., 2015). The first 4 volumes of echo-planar images were excluded, and the skull was stripped to reduce head movement. Images were motion corrected, despiked, and normalized to a grand-mean of 10,000. Data were band-pass filtered to frequencies between 0.01 Hz and 0.1 Hz and detrended using first- and second-order polynomials.
Images were spatially smoothed using a gaussian kernel (full width at the half maximum = 8 mm). They were registered to standard space using the Montreal Neurological Institute template. We excluded volumes in which the framewise displacement (Yan et al., 2013) or the temporal derivative of the RMS variance over the voxels (Power et al., 2012) was larger than the 95% percentile of the total sample. We regressed out the following nuisance covariates: cerebral spinal fluid, white matter, global signal, and 6 linear motion parameters.
The mean blood oxygenation level dependent signal of each region of interest was extracted using spheres with a 5-mm radius, and the scrubbing procedure was conducted in R platform for computational statistics (www.r-project.org). The pairwise functional connectivity weights were obtained using the Spearman correlation coefficient also in R. We calculated the connectivity among the main nodes from each of the 10 tau pathology networks previously described (Hoenig et al., 2018), which included the left temporal middle gyrus, left precentral gyrus, right precuneus, right superior occipital region, left fusiform gyrus, left posterior cingulate cortex, right cuneus, right frontal medial orbital region, right superior temporal gyrus, and left parahippocampal gyrus.
2.4. Cognitive function
2.4.1. Nondeclarative memory
Nondeclarative memory was assessed using the Rey-Osterrieth Complex Figure Test (ROCFT). Participants were asked to draw the ROCFT figure in 3 steps: while looking at it (copy) and from memory, after 3 and 30 minutes (immediate and delayed recall, respectively). The ROCFT scores were calculated using the mean percent-retained items for each item (i.e., recall score/copy score), as previously described (Axelrud et al., 2018). Dependent variables were factor scores for each task after regressing out age trends by saving studentized residuals. Previous research reported associations between the AD-PRS and impaired performance in ROCFT in children and adolescents (Axelrud et al., 2018).
2.4.2. Executive function
Executive function was evaluated using tasks that investigated working memory, inhibitory control, and time processing. Working memory was assessed using the backward digital span (Wechsler et al., 2002), in which individuals were asked to repeat sequences of numbers in the order stated or in the reverse order; and backward Corsi blocks (Wechsler et al., 2002), in which participants were asked to repeat a spatial sequence tapped in 9 identical blocks. Inhibitory control was assessed using the conflict control task (Vandierendonck et al., 2004), which involved indicating the direction of an arrow and the go/no go task (Hogan et al., 2005) in which individuals had to inhibit the tendency to press a button indicating the directing of arrows when a double-headed arrow appeared. Time-processing task was investigated with a task in which subjects had to anticipate the appearance of a visual stimulus after 400 ms and 2,000 ms (Bitsakou et al., 2008). All indexes were combined into a global executive function score using confirmatory factor analysis (Martel et al., 2017). Dependent variables were factor scores for each task after regressing out age trends by saving studentized residuals.
2.5. Statistical analyses
We selected spheres with a 5-mm radius centered at coordinates reported previously (Hoenig et al., 2018). The correlation of the blood oxygenation level dependent signal between regions was calculated using the Pearson coefficient, creating a matrix with 45 correlations. We used Student t-tests against 0 to determine if brain regions were functionally connected in our samples. Only nodes functionally connected (p < 0.05, adjusted for multiple comparisons using the Benjamini-Hochberg method) were considered for further analyses.
Main analyses were tested with multiple regression models using connections between the main node of each tau pathology network as dependent variables and the AD-PRS as an independent variable. We used as covariates 4 principal components from genotyping, to adjust for genetic population stratification, and mean framewise displacement after scrubbing, to minimize head movement bias. The 4 principal components from genotyping were derived both separately for each sample (for the discovery/replication analyses) and considering both samples together (for the exploratory analyses). We also calculated ancestry using the Admixture software (Alexander et al., 2009). All analyses considered sampling weights, which adjust for our high-risk selection procedure (Martel et al., 2017). We used a discovery/replication approach, arbitrarily choosing the Porto Alegre site as the discovery sample, as in our previous study (Axelrud et al., 2018). All analyses in the discovery sample were adjusted for multiple comparisons using the Benjamini-Hochberg method. Significant associations in the discovery sample were then tested in the replication sample (São Paulo site), using a threshold of p < 0.05.
For the connections significantly associated to the AD-PRS, we also conducted 4 sensitivity analyses (i.e., investigating how strong the findings are allowing for variation in methodological decisions), 2 exploratory analyses (i.e., assessing the meaning of the findings in terms of association with clinically relevant phenotypes), and 2 specificity analyses (i.e., assessing whether results were specific to the AD-PRS and not found in PRS for other disorders).
Sensitivity analyses included (1) adjusted analysis for sex and age, APOE genotype, European ancestry scores, and 10 principal components from genotyping (instead of 4); (2) selecting only Caucasian subjects (n = 374); (3) assessing the impact of distinct p-thresholds of the AD-PRS; and (4) evaluating associations with the APOE genotype. Exploratory analyses were performed in the total sample and included (1) regressions investigating associations of the AD-PRS related connections with executive function and its 3 domains (adjusting for multiple testing) and nondeclarative memory and (2) an analysis examining whether these connections moderated the association between the AD-PRS and nondeclarative memory. For the latest analysis, we used marginal effects estimation, which represents the effects on predicted levels of memory recall for one standardized unit change of the AD-PRS or the connectivity of the significant tau pathology networks, when the other predictor (connectivity or AD-PRS) is held constant at different values (−2.0 to 2.0 standard deviations [SDs]). The interaction was graphically represented using R packages “interplot” (Solt and Hu, 2015) and “persp3D” (Soetaert, 2016), and marginal effects were explored using STATA, version 13 (StataCorp, College Station, TX). Specificity analyses included regressions between the AD-PRS–related connections and the PRS for schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and major depression (Wray et al., 2018), which were calculated similarly to the AD-PRS. We selected a p-threshold lower than 0.05 (nSNP schizophrenia: 13,092; nSNP depression: 9549) based on previous studies (Musliner et al., 2019).
3. Results
3.1. Participants
Participants’ description can be found in Table 1. Compared with the original sample (N = 2512), individuals who underwent genotyping had a similar mean age (original sample: mean = 124.6 months, SD = 23.05; genotyped sample: mean = 121.2 months, SD = 22.31) and sex distribution (original sample: 46.7% female; genotyped sample: 45.9% female). The AD-PRS was significantly lower in the discovery sample than in the replication sample (F = 21.5, p < 0.001). Also, the AD-PRS was not associated with age (B = −0.017, p = 0.571), gender (F = 0.162, p = 0.688), and family income (B = −0.08, p = 0.141).
Table 1.
Samples description
| Variables | Discovery sample (n = 332) | Replication sample (n = 304) |
|---|---|---|
| Mean (SD) | ||
| Age | 10.96 (1.97) | 10.64 (1.73) |
| Monthly family income (USD) | 768.23 (601.24) | 964.21 (614.48) |
| Mean AD-PRS | 0.00155 (0.00046) | 0.00169 (0.00041) |
| N (%) | ||
| Female | 164 (52.10) | 175 (59.10) |
| Ethnicity (self-declared) | ||
| Caucasian | 214 (66.30) | 160 (54.10) |
| Black | 46 (14.20) | 18 (6.10) |
| Multiracial | 59 (18.30) | 116 (39.20) |
| Indigenous | 3 (0.90) | 1 (0.30) |
| Asian | 0 (0) | 1 (0.30) |
| Number of APOE E4 alleles | ||
| 0 | 255 (76.80) | 227 (74.70) |
| 1 | 67 (20.20) | 67 (22.00) |
| 2 | 10 (3.00) | 7 (2.30) |
| Unknown | - | 3 (1.00) |
Key: AD-PRS, polygenic risk score for Alzheimer’s disease; APOE, apolipoprotein E; SD, standard deviation.
3.2. Main analyses
Student t-test analyses showed 35 connections were significantly different from 0 in the discovery and replication samples (see Supplemental Material).
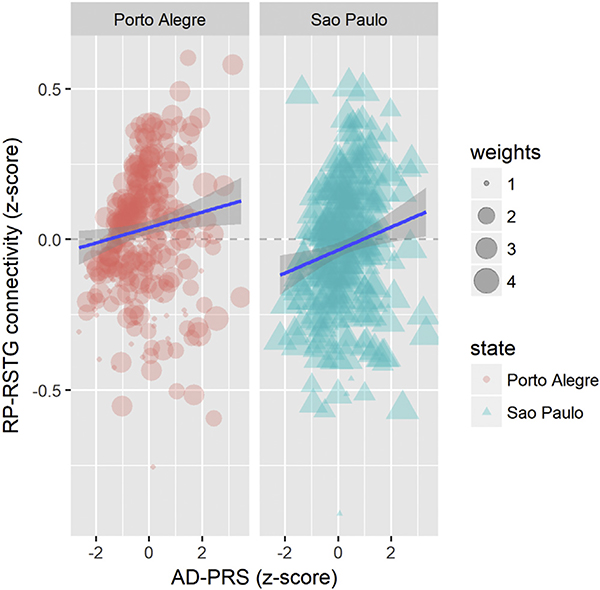
Regressions revealed a one-unit increase in the AD-PRS z-score corresponded to a 0.18 z-score increment in the connectivity between the right precuneus and the right superior temporal gyrus (pcrude = 0.005, padjusted = 0.036). Similar results were obtained for the replication sample (β = 0.202, p = 0.031) (Fig. 1). We also found associations that reached trend-level replications between the AD-PRS and the connectivity between the right superior temporal gyrus and the left middle temporal gyrus (β= −0.198, padjusted = 0.017 for the discovery sample, β = −0.176, p = 0.051 for the replication sample). No other replicable associations of the AD-PRS with connections of tau pathology networks emerged from this analysis (see Supplemental Material).
Fig. 1.
Associations of the polygenic risk score for Alzheimer’s disease (AD-PRS) with the connectivity between the right precuneus and the right superior temporal gyrus (RP-RSTG) for the discovery and replication samples. Note: Weights represent sampling weights, as explained in Section 2.
3.3. Sensitivity analyses
The association between the AD-PRS and the connectivity between the right precuneus and the right superior temporal gyrus remained significant when adjusting for age and sex, European ancestry, APOE genotype, and for 10 principal components (see Supplemental Material). Results were also significant for a Caucasian subsample (β = 0.179, p = 0.004). Furthermore, associations between the AD-PRS and the connectivity between the right precuneus and the right superior temporal gyrus were significant when using the AD-PRS calculated using the stringent thresholds 0.05 to 0.2 and trend level using p-thresholds lower than 0.3 to 0.5 in the discovery sample (see Supplemental Material). We did not find an association of this connectivity with the APOE genotype (F = 0.046, p = 0.830).
3.4. Exploratory analysis
We did not find an association of the connectivity between the right precuneus and the right superior temporal gyrus with global executive function (β = 0.064, p = 0.120). Regarding executive function domains, we found an association between this connectivity and inhibitory control (β = 0.157, padjusted = 0.035), but not working memory (β = 0.049, padjusted = 0.222) and time processing (β = 0.055, padjusted = 0.222).
We also did not find associations of the connectivity between the right precuneus and the right superior temporal gyrus with immediate (β = −0.025, p = 0.537) and delayed recall (β = −0.031, p = 0.426). Nevertheless, we found this connectivity moderated the associations of AD-PRS with both immediate (p = 0.004) and delayed recall (p = 0.016), which were described in a previous study (Axelrud et al., 2018).
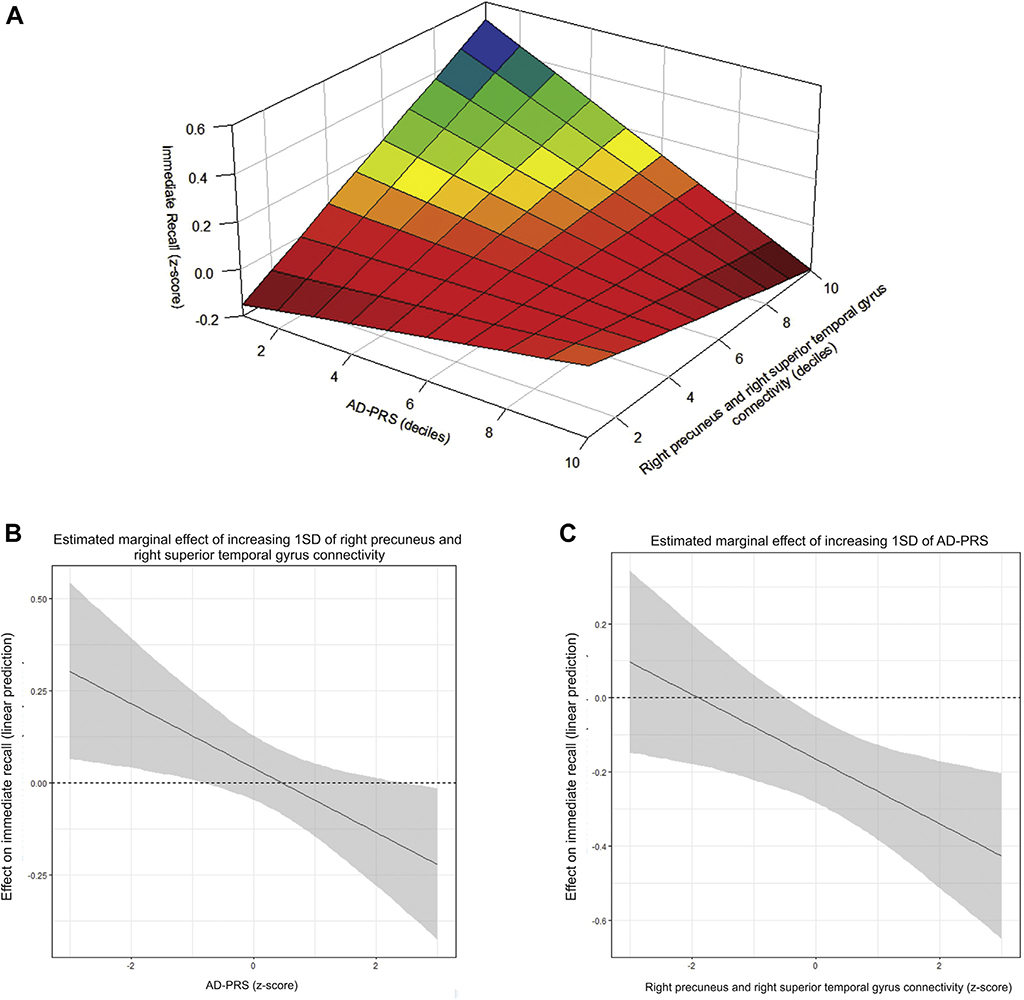
Marginal effect analysis revealed that, for subjects with high levels of genetic susceptibility to AD (≥1.5 z-score), the connectivity between the right precuneus and the right superior temporal gyrus was negatively associated with immediate recall, whereas for subjects with low genetic susceptibility to AD (≤−0.5 z-score), this association was positive (Table 2 and Fig. 2, Panel B). Furthermore, the AD-PRS was significantly associated with lower performance in immediate recall only for subjects in which the connectivity between the right precuneus and the right superior temporal gyrus was higher than average (representation in Fig. 2, Panel C). The effect of both the AD-PRS and the connectivity between the right precuneus and the right superior temporal gyrus quantiles on immediate recall z-score is represented in Fig. 2, Panel A. Similar results were found for delayed recall (Table 3).
Table 2.
Marginal effects of increasing one standard deviation of the RP-RSTG or the AD-PRS for fixed values of each predictor on immediate recall performance
| AD-PRS z-score | RP-RSTG |
AD-PRS |
|
|---|---|---|---|
| β (95% CI) | RP-RSTG z-score | β (95% CI) | |
| −2.0 | 0.356*** (0.219 to 0.494) | −2.0 | 0.113** (0.036 to 0.190) |
| −1.5 | 0.289*** (0.163 to 0.416) | −1.5 | 0.046 (0.036 to 0.190) |
| −1.0 | 0.223*** (0.108 to 0.416) | −1.0 | −0.020 (−0.120 to 0.080) |
| −0.5 | 0.156** (0.054 to 0.259) | −0.5 | −0.087 (−0.198 to 0.025) |
| 0.0 | 0.089 (−0.002 to 0.181) | 0.0 | −0.153* (−0.277 to −0.030) |
| 0.5 | 0.023 (−0.057 to 0.103) | 0.5 | −0.220** (−0.355 to −0.085) |
| 1.0 | −0.043 (−0.057 to 0.103) | 1.0 | −0.286*** (−0.433 to −0.140) |
| 1.5 | −0.110*** (−0.167 to 0.053) | 1.5 | −0.353*** (−0.511 to −0.195) |
| 2.0 | −0.177*** (−0.222 to 0.131) | 2.0 | −0.420*** (−0.589 to −0.250) |
Marginal effects derived from interaction models of PRS with RP-RSTG connectivity for immediate recall test.
Key: β, regression coefficient predicting immediate recall; AD-PRS, polygenic risk score for Alzheimer’s disease; CI, confidence interval; RP-RSTG, right precuneus and the right superior temporal gyrus.
p < 0.001
p < 0.01
p < 0.05.
Fig. 2.
Interaction and marginal effects of the connectivity between the right precuneus and the right superior temporal gyrus and the AD-PRS on immediate recall. Note: Panel A depicts the immediate recall test performance (z-scores) according to deciles of the AD-PRS and the right precuneus and the right superior temporal gyrus connectivity. Average marginal effects for the linear prediction of immediate recall (y-axis) are depicted in Panels B and C. In Panel B, the estimated marginal effect of increasing levels (1 SD) of the right precuneus and the right superior temporal gyrus connectivity are demonstrated by each z-score of AD-PRS (x-axis). In Panel C, the estimated marginal effect of increasing levels (1 SD) of the AD-PRS is demonstrated by each z-score of the right precuneus and the right superior temporal gyrus connectivity (x-axis). Abbreviations: SD, standard deviation; AD-PRS, polygenic risk score for Alzheimer’s disease.
Table 3.
Marginal effects of increasing one standard deviation of the RP-RSTG or the AD-PRS for fixed values of each predictor on delayed recall performance
| AD-PRS z-score | RP-RSTG |
AD-PRS |
|
|---|---|---|---|
| β (95% CI) | RP-RSTG z-score | β (95% CI) | |
| −2.0 | 0.318*** (0.273 to 0.364) | −2.0 | 0.082 (−0.097 to 0.260) |
| −1.5 | 0.259 *** (0.229 to 0.289) | −1.5 | 0.022 (−0.141 to 0.186) |
| −1.0 | 0.200*** (0.185 to 0.214) | −1.0 | −0.036 (−0.185 to 0.111) |
| −0.5 | 0.141*** (0.140 to 0.141) | −0.5 | −0.096 (−0.229 to 0.036) |
| 0.0 | 0.081*** (0.065 to 0.098) | 0.0 | −0.155** (−0.273 to −0.038) |
| 0.5 | 0.022 (−0.009 to 0.054) | 0.5 | −0.215*** (−0.316 to −0.113) |
| 1.0 | −0.037 (−0.084 to 0.010) | 1.0 | −0.274*** (−0.360 to −0.187) |
| 1.5 | −0.096** (−0.159 to −0.034) | 1.5 | −0.333*** (−0.404 to −0.262) |
| 2.0 | −0.156*** (−0.233 to −0.078) | 2.0 | −0.392*** (−0.448 to −0.337) |
Marginal effects derived from interaction models of PRS with RP-RSTG connectivity for immediate recall test.
Key: β, regression coefficient predicting immediate recall; AD-PRS, polygenic risk score for Alzheimer’s disease; CI, confidence interval; RP-RSTG, right precuneus and the right superior temporal gyrus.
p < 0.001
p < 0.01
p < 0.05.
3.5. Specificity analyses
We did not find associations of the connection between the right precuneus and the right superior temporal gyrus with the PRS for schizophrenia (β = −0.038, p = 0.707) and depression (β = 0.008, p = 0.955).
4. Discussion
We found that the AD-PRS, but not APOE, was associated with an increased connectivity between the right precuneus and the right superior temporal gyrus. These regions are the local maxima of tau pathology networks that most likely represent the ventral default mode network and the language network (Hoenig et al., 2018), which are known to be impaired in AD (Weiler et al., 2014). Furthermore, we found this connectivity is associated with inhibitory control and moderates the association of the AD-PRS with both immediate and delayed recall. These findings suggest the connectivity between regions susceptible to tau pathology in late life may be affected in early life in individuals with genetic susceptibility to AD, which might impact both inhibitory control and memory.
The stereotypical anatomical propagation of tau pathology in AD might result from the network degeneration hypothesis, in which misfolded tau proteins would spread along functional networks (Hoenig et al., 2018; Seeley et al., 2009). Here, we showed connections between regions associated with the ventral default mode network and language networks are already aberrant in children and adolescents with higher genetic susceptibility to AD. Abnormalities in this connection were also revealed in a previous study, which showed significant, but not replicable, associations between higher levels of p-tau and increased connectivity between these areas (Tucholka et al., 2017). Our findings are also supported by previous studies, which have shown reduced volume and altered metabolism in the precuneus and in temporal regions in cognitively normal adults with a family history of AD (Mosconi et al., 2014) and in infant ε4 carriers (Dean et al., 2014). In keeping with the literature (Axelrud et al., 2018; Su et al., 2017), our findings suggest brain connectivity might be affected by genetic risk for AD in childhood and adolescence and predispose individuals to this disease later in life contingent on other genetic factors and environmental influences.
Furthermore, previous research showed associations of the AD-PRS with memory and hippocampal volume in youth (Axelrud et al., 2018). Our findings extend past evidence showing the association between the AD-PRS and memory is only significant in subjects with a strong connectivity between the right precuneus and the right superior temporal gyrus. This provides further insights on AD pathogenesis and on implications of genetic susceptibility to AD development in the brain during childhood and adolescence. Future studies can examine biological mechanisms that could explain this interaction and investigate whether this connectivity could help understand pathways leading to susceptibility to dementia in older individuals with high genetic risk to AD.
Limitations of this research need to be addressed. First, the use of multiple comparisons might increase the risk of type 1 error. Nevertheless, the correction for multiple comparisons in the main analyses and the replication in a second sample decrease this possibility. Second, analyses were performed cross-sectionally, and therefore, it is not possible to determine whether this connectivity would still be associated with AD risk later in life. Future studies could investigate the implications of the AD-PRS in tau pathology networks over the lifespan. Third, given that the AD-PRS is not specific to tau pathology and that few tau abnormalities are present in such young ages, different neurological pathways, such as β amyloid accumulation and inflammation, might be contributing to our results and limiting their specificity. This nonspecificity might also explain the lack of association with the APOE gene.
Our results suggest brain connectivity may be affected during childhood and adolescence in individuals with genetic susceptibility to AD, which might impact memory performance and inhibitory control. Further research is necessary to replicate those findings and to continue advancing on the impact of genetic susceptibility to AD in early life.
Supplementary Material
Acknowledgements
The authors thank the children and families from the Brazilian High-Risk Study for their participation, which made this research possible. The authors also thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses.
Disclosures
This work was funded through research grants by the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq, Brazil; grant number 573974/2008-0), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil, the Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil (FAPESP, Brazil; grant number 2008/57896-8), and the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul, Brazil. All of them are public institutions of the Brazilian government developed for scientific research support. Dr. Pine’s efforts were supported by the National Institute of Mental Health Intramural Research Program. Funding sources have no involvement in this study, including no role in data collection, analysis, and interpretation of the data.
Ms. Axelrud, Dr. Sato, Dr. Santoro, Ms. Fernanda Talarico, Dr. Pine, Dr. Zugman, Dr. Amaro, Dr. Grassi-Oliveira, Dr. Guinjoan, Dr. Simioni, Dr. Hoffmann, Dr. Hakonarson, Dr. Brietzke, Dr. Gadelha, Dr. Da Silva, Dr. Hoexter, Dr. Belangero, Dr. Miguel, and Dr. Salum report no biomedical financial interests or potential conflicts of interest. Dr. Pan has received payment for the development of educational material for Janssen-Cilag and AstraZeneca. Dr. Bressan has been on the speakers’ bureau/advisory board of AstraZeneca, Bristol, Janssen, and Lundbeck; he has received research grants from Janssen, Eli-Lilly, Lundbeck, Novartis, Roche, FAPESP, CNPq, CAPES, Fundação E.J. Safra, and Fundação ABAHDS. He is a shareholder of Biomolecular Technology Ltda. Dr. Rohde has received Honoraria, has been on the speakers’ bureau/advisory board, and/or has acted as a consultant for Eli-Lilly, Janssen-Cilag, Novartis, and Shire in the last two years. He receives authorship royalties from Oxford Press and ArtMed. He also received travel awards for taking part of 2014 APA and 2015 WFADHD meetings from Shire. The ADHD and Juvenile Bipolar Disorder Outpatient Programs chaired by him received unrestricted educational and research support from the following pharmaceutical companies in the last three years: Eli-Lilly, Janssen-Cilag, Novartis, and Shire.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neurobiolaging.2019.06.011.
References
- Alexander DH, Novembre J, Lange K, 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrud LK, Santoro ML, Pine DS, Talarico F, Gadelha A, Manfro GG, Pan PM, Jackowski A, Picon F, Brietzke E, Grassi-Oliveira R, Bressan RA, Miguel EC, Rohde LA, Hakonarson H, Pausova Z, Belangero S, Paus T, Salum GA, 2018. Polygenic risk score for Alzheimer’s disease: implications for memory performance and hippocampal volumes in early life. Am. J. Psychiatry 175, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badhwar A, Tam A, Dansereau C, Orban P, Hoffstaedter F, Bellec P, 2017. Resting-state network dysfunction in Alzheimer’s disease: a systematic review and meta-analysis. Alzheimers Dement. (Amst.) 8, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitsakou P, Psychogiou L, Thompson M, Sonuga-Barke EJS, 2008. Inhibitory deficits in attention-deficit/hyperactivity disorder are independent of basic processing efficiency and IQ. J. Neural Transm 115, 261–268. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, 2011. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 121, 171–181. [DOI] [PubMed] [Google Scholar]
- Braak H, Thal DR, Ghebremedhin E, Del Tredici K, 2011. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J. Neuropathol. Exp. Neurol 70, 960–969. [DOI] [PubMed] [Google Scholar]
- Dean DC 3rd, Jerskey BA, Chen K, Protas H, Thiyyagura P, Roontiva A, O’Muircheartaigh J, Dirks H, Waskiewicz N, Lehman K, Siniard AL, Turk MN, Hua X, Madsen SK, Thompson PM, Fleisher AS, Huentelman MJ, Deoni SCL, Reiman EM, 2014. Brain differences in infants at differential genetic risk for late-onset Alzheimer disease: a cross-sectional imaging study. JAMA Neurol 71, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elman JA, Madison CM, Baker SL, Vogel JW, Marks SM, Crowley S, O’Neil JP, Jagust WJ, 2016. Effects of beta-amyloid on resting state functional connectivity within and between networks reflect known patterns of regional vulnerability. Cereb. Cortex 26, 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF, 2015. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson O, Grothe MJ, Strandberg TO, Ohlsson T, Hagerstrom D, Jogi J, Smith R, Scholl M, 2017. Tau pathology distribution in Alzheimer’s disease corresponds differentially to cognition-relevant functional brain networks. Front. Neurosci 11, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenig MC, Bischof GN, Seemiller J, Hammes J, Kukolja J, Onur OA, Jessen F, Fliessbach K, Neumaier B, Fink GR, van Eimeren T, Drzezga A, 2018. Networks of tau distribution in Alzheimer’s disease. Brain 141, 568–581. [DOI] [PubMed] [Google Scholar]
- Hogan AM, Vargha-Khadem F, Kirkham FJ, Baldeweg T, 2005. Maturation of action monitoring from adolescence to adulthood: an ERP study. Dev. Sci 8, 525–534. [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin C-F, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau M-T, Choi S-H, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, BarbergerGateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Zompo MD, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RFAG, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JSK, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang L, Dartigues J-F, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z-L, Tan Y-F, Qiu Y-J, Zhu J-P, Chen Y, Lin S-S, Wu M-H, Mao Y-P, Hu J-J, Ding Z-X, Yu E-Y, 2018. Interhemispheric functional connectivity for Alzheimer’s disease and amnestic mild cognitive impairment based on the triple network model. J. Zhejiang Univ. Sci. B 19, 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel MM, Pan PM, Hoffmann MS, Gadelha A, do Rosario MC, Mari JJ, Manfro GG, Miguel EC, Paus T, Bressan RA, Rohde LA, Salum GA, 2017. A general psychopathology factor (P factor) in children: structural model analysis and external validation through familial risk and child global executive function. J. Abnorm. Psychol 126, 137–148. [DOI] [PubMed] [Google Scholar]
- Mormino EC, Sperling RA, Holmes AJ, Buckner RL, De Jager PL, Smoller JW, Sabuncu MR, 2016. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology 87, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Murray J, Tsui WH, Li Y, Spector N, Goldowsky A, Williams S, Osorio R, McHugh P, Glodzik L, Vallabhajosula S, de Leon MJ, 2014. Brain imaging of cognitively normal individuals with 2 parents affected by late-onset AD. Neurology 82, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musliner KL, Mortensen PB, McGrath JJ, Suppli NP, Hougaard DM, Bybjerg-Grauholm J, Bækvad-Hansen M, Andreassen O, Pedersen CB, Pedersen MG, Mors O, Nordentoft M, Børglum AD, Werge T, Agerbo E, Consortium, for the B.D.W.G. of the P.G, 2019. Association of polygenic liabilities for major depression, bipolar disorder, and schizophrenia with risk for depression in the Danish PopulationAssociation of polygenic liabilities with risk for depression in the Danish PopulationAssociation of polygenic liabilities with risk for depression in the Danish population. JAMA Psychiatry 76, 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg A, 2015. Dementia in 2014. Towards early diagnosis in Alzheimer disease. Nat. Rev. Neurol 11, 69–70. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Mucke L, 2006. A network dysfunction perspective on neurodegenerative diseases. Nature 443, 768–773. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE, 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salum GA, Gadelha A, Pan PM, Moriyama TS, Graeff-Martins AS, Tamanaha AC, Alvarenga P, Valle Krieger F, Fleitlich-Bilyk B, Jackowski A, Sato JR, Brietzke E, Polanczyk GV, Brentani H, de Jesus Mari J, Do Rosario MC, Manfro GG, Bressan RA, Mercadante MT, Miguel EC, Rohde LA, 2015. High risk cohort study for psychiatric disorders in childhood: rationale, design, methods and preliminary results. Int. J. Methods Psychiatr. Res 24, 58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, Salum GA, Gadelha A, Vieira G, Zugman A, Picon FA, Pan PM, Hoexter MQ, Anes M, Moura LM, Del’Aquilla MAG, Crossley N, Amaro Junior E, Mcguire P, Lacerda ALT, Rohde LA, Miguel EC, Jackowski AP, Bressan RA, 2015. Decreased centrality of subcortical regions during the transition to adolescence: a functional connectivity study. Neuroimage 104, 44–51. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD, 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron 62, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetaert K, 2016. plot3D: Plotting Multi-Dimensional Data. http://CRAN.R-project.org/package=plot3D. (Accessed 26 April 2019).
- Solt F, Hu Y, 2015. interplot: Plot the effects of variables in interaction terms. http://CRAN.R-project.org/package=interplot. (Accessed 26 April 2019).
- Su YY, Zhang XD, Schoepf UJ, Varga-Szemes A, Stubenrauch A, Liang X, Zheng LJ, Zheng G, Kong X, Xu Q, Wang SJ, Qi RF, Lu GM, Zhang LJ, 2017. Lower functional connectivity of default mode network in cognitively normal young adults with mutation of APP, presenilins and APOE epsilon4. Brain Imaging Behav 11, 818–828. [DOI] [PubMed] [Google Scholar]
- Tucholka A, Grau-Rivera O, Falcon C, Rami L, Sánchez-Valle R, Lladó A, Gispert JD, Molinuevo JL, Alzheimer’s Disease Neuroimaging Initiative, 2017. Structural connectivity alterations along the Alzheimer’s disease continuum: reproducibility across two independent samples and correlation with cerebrospinal fluid amyloid-b and tau. J. Alzheimers Dis 61, 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandierendonck A, Kemps E, Fastame MC, Szmalec A, 2004. Working memory components of the Corsi blocks task. Br. J. Psychol 95, 57–79. [DOI] [PubMed] [Google Scholar]
- Wang L, Roe CM, Snyder AZ, Brier MR, Thomas JB, Xiong C, Benzinger TL, Morris JC, Ances BM, 2012. Alzheimer disease family history impacts resting state functional connectivity. Ann. Neurol 72, 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhou B, Yao H, Zhan Y, Zhang Z, Cui Y, Xu K, Ma J, Wang L, An N, Zhang X, Liu Y, Jiang T, 2015. Aberrant intra- and inter-network connectivity architectures in Alzheimer’s disease andmild cognitive impairment. Sci. Rep 5,14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D, Simões M, Ferreira C, 2002. WISC-III: escala de inteligência de Wechsler para crianças III. Casa do Psicólogo, São Paulo. [Google Scholar]
- Weiler M, Fukuda A, Massabki LHP, Lopes TM, Franco AR, Damasceno BP, Cendes F, Balthazar MLF, 2014. Default mode, executive function, and language functional connectivity networks are compromised in mild Alzheimer’s disease. Curr. Alzheimer Res 11, 274–282. [DOI] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu S-A, Bækvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschøn HN, Bybjerg-Grauholm J, Cai N, Castelao E, Christensen JH, Clarke T-K, Coleman JIR, Colodro-Conde L, CouvyDuchesne B, Craddock N, Crawford GE, Crowley CA, Dashti HS, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV, Dunn EC, Eley TC, Eriksson N, Escott-Price V, Kiadeh FHF, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Giusti-Rodríguez P, Goes FS, Gordon SD, Grove J, Hall LS, Hannon E, Hansen CS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga J-J, Hougaard DM, Hu M, Hyde CL, Ising M, Jansen R, Jin F, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Lane JM, Li Y, Li Y, Lind PA, Liu X, Lu L, MacIntyre DJ, MacKinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, McGrath P, McGuffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mill J, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B, Nivard MG, Nyholt DR, O’Reilly PF, Oskarsson H, Owen MJ, Painter JN, Pedersen CB, Pedersen MG, Peterson RE, Pettersson E, Peyrot WJ, Pistis G, Posthuma D, Purcell SM, Quiroz JA, Qvist P, Rice JP, Riley BP, Rivera M, Saeed Mirza S, Saxena R, Schoevers R, Schulte EC, Shen L, Shi J, Shyn SI, Sigurdsson E, Sinnamon GBC, Smit JH, Smith DJ, Stefansson H, Steinberg S, Stockmeier CA, Streit F, Strohmaier J, Tansey KE, Teismann H, Teumer A, Thompson W, Thomson PA, Thorgeirsson TE, Tian C, Traylor M, Treutlein J, Trubetskoy V, Uitterlinden AG, Umbricht D, Van der Auwera S, van Hemert AM, Viktorin A, Visscher PM, Wang Y, Webb BT, Weinsheimer SM, Wellmann J, Willemsen G, Witt SH, Wu Y, Xi HS, Yang J, Zhang F, Arolt V, Baune BT, Berger K, Boomsma DI, Cichon S, Dannlowski U, de Geus ECJ, DePaulo JR, Domenici E, Domschke K, Esko T, Grabe HJ, Hamilton SP, Hayward C, Heath AC, Hinds DA, Kendler KS, Kloiber S, Lewis G, Li QS, Lucae S, Madden PFA, Magnusson PK, Martin NG, McIntosh AM, Metspalu A, Mors O, Mortensen PB, Müller-Myhsok B, Nordentoft M, Nöthen MM, O’Donovan MC, Paciga SA, Pedersen NL, Penninx BWJH, Perlis RH, Porteous DJ, Potash JB, Preisig M, Rietschel M, Schaefer C, Schulze TG, Smoller JW, Stefansson K, Tiemeier H, Uher R, Völzke H, Weissman MM, Werge T, Winslow AR, Lewis CM, Levinson DF, Breen G, Børglum AD, Sullivan PF, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, 2018. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C-G, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo X-N, Castellanos FX, Milham MP, 2013. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage 76, 183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.