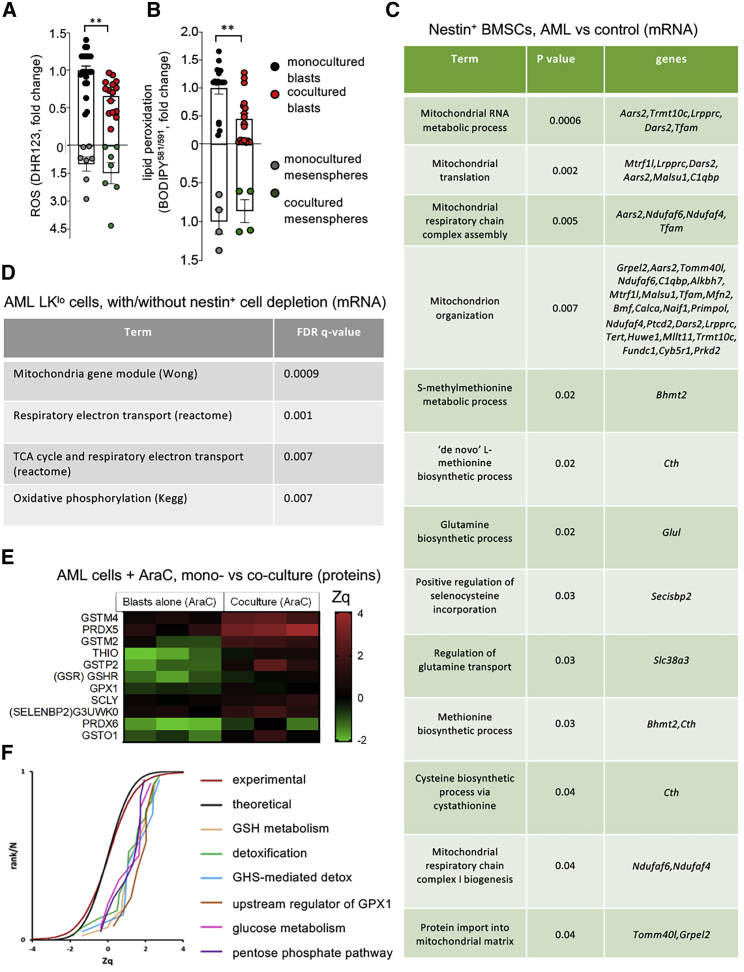
Figure 4.
BMSCs Provide Leukemic Blasts with Antioxidant Defense during Chemotherapy
(A and B) ROS measured by DH123 staining (A) and lipid peroxidation detected with BODIPY581/591 (B) in AML blasts and BMSCs cultured alone or together for 24 h in presence of AraC. Each dot is a biological replicate. ∗∗p < 0.05; unpaired two-tailed t test.
(C) Selected pathways from the GO enrichment analysis of significantly upregulated mRNA (RNA-seq) of CD45−CD31−Ter119−Nes-GFP+ cells isolated from the BM of primary iMLL-AF9 mice or control mice. The GO term, adjusted p value and differentially expressed genes in each category are indicated.
(D) Selected pathways from the GSEA analysis of RNA-seq of hematopoietic-lineage-negative ckitlow (LKlo) cells isolated from the BM of AML mice with or without nestin+ cell depletion. Lethally irradiated CD45.2 control mice or Nes-creERT2;iDTA mice were transplanted with 106 iAML (rtTA;MLL-AF9) CD45.2+ BM nucleated cells and 106 CD45.1+ WT BM nucleated cells. Doxycycline administration started 2 weeks after transplant; tamoxifen was administered 4 weeks post-transplant; and mice were sacrificed and analyzed 4 weeks later. BM MLL-AF9+ lin− ckitlow (LKlo) cells were sorted from leukemic mice with or without nestin+ cell depletion. The pathway term, signature, and adjusted p value are indicated.
(E) Heatmap showing antioxidant and GSH related among the top upregulated (red) protein pathways detected by proteomics in AraC-treated AML blasts cultured alone or cocultured with mesenspheres for 24 h. Quantitative proteomics results were analyzed using the WSPP statistical model (Navarro et al., 2014) and expressed in log2 fold change values in units of SD (Zq) with respect to baseline. Significant protein changes (adjusted p < 0.05) are indicated; two-sided Fisher test, 3 biological replicates in each condition.
(F) SBT analysis (García-Marqués et al., 2016) to detect changes in functional categories due to coordinated protein behavior. The analysis detected significantly increased pathways (positive values of Zq indicate increased protein changes) compared with the theoretical N(0,1) distribution (black curve); for comparison, the distribution of Zq values of all the proteome is also shown (red curve). The panel displays the cumulative distributions of Zq from proteins belonging to a set of representative altered categories, showing the high coordination of protein responses in these pathways.