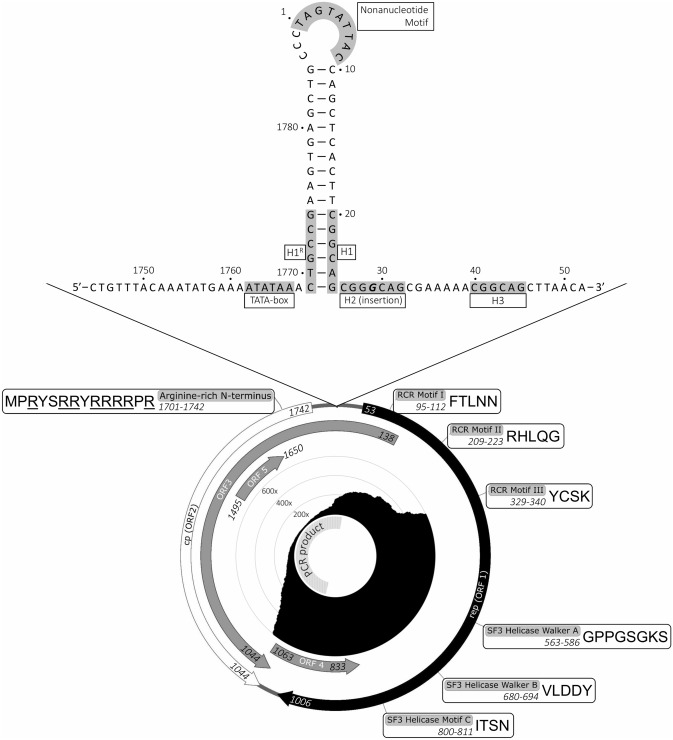
Figure 1.
Structure of the elk circovirus Banff/2019 genome. Bottom: Complete circular genome map and coverage plot. On the outside, start and end nucleotide position of ORFs and motifs are indicated in italics. Amino acid sequences (5′–3′) are shown in single letter code beside motif names and positions. ORFs were visualized using SnapGene Viewer v5.0.7 (snapgene.com). Inside the annotated circular genome is a circular plot (generated using GView v1.738) showing coverage at each nucleotide position based on reference mapping of the liver-derived metagenomic sequencing data to the assembled genome. Inside the coverage plot, an 874 nt PCR product (from nucleotide positon 963 to 49) that was subsequently sequenced in order to confirm a region with low relative coverage, is indicated (coverage data not shown). Top: Expanded view of the origin of replication stem-loop region. Nucleotide position and features of note are labelled. Hexamer repeats are labelled as H1-3 with a reverse orientation repeat on the opposite stem of the stem-loop compared to H1 labelled as H1R. A single base insertion located in the H2 hexamer repeat sequence that is also found in BatACV11 is indicated in bold and italics.