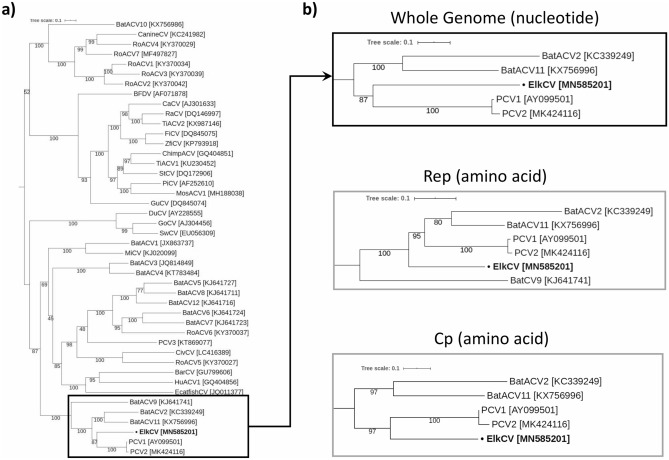
Figure 2.
Maximum likelihood phylogenetic trees of representative sequences within the genus Circovirus. Sequences were aligned using MAFFT24, trees generated using IQ-Tree23 on find best model setting with ModelFinder25 with 1000 ultrafast bootstraps26 and visualized with iTOL27. ElkCV is indicated in bold text and with “•” before the sequence name in all trees. (a) Full mid-point rooted phylogenetic tree generated from representative Circovirus complete genome nucleotide sequences (model TIM + F + R5). (b) Pruned trees showing only ElkCV and sequences on closely related branches from complete genome nucleotide sequences (top black box), Rep protein amino acid sequences (middle gray box, model LG + I + G4) and Cp protein amino acid sequences (bottom gray box, model PMB + R3).