Figure 3.
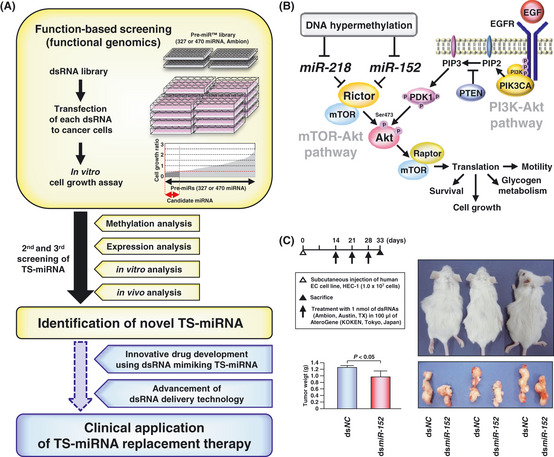
Function‐based screening of tumor‐suppressive micro RNA (TS‐miRNA) for miRNA replacement therapy as a cancer treatment. (A) Strategy of our function‐based approach to the identification of epigenetically silenced TS‐miRNA in cancer cells. Previously, to identify novel TS‐miRNA having the great potential for miRNA replacement therapy, we performed function‐based screening combined with methylation and expression analyses in oral squamous cell carcinoma (OSCC) and endometrial cancer (EC) cell lines according to this strategy shown in this figure, resulting in identification of novel TS‐miRNA,miR‐218 and ‐152, respectively, directly targeting Rictor.23, 24 (B) A model summarizing the molecular mechanism of miR‐218,miR‐152 and their direct target Rictor in the TOR‐Akt signaling pathway. Our previous studies demonstrated that these TS‐miRNA acted as suppressors of the TOR‐Akt signaling pathway, independently of the PI3K‐Akt signaling pathway and that methylation‐mediated silencing of these TS‐miRNA might contribute to the pathogenesis of OSCC and EC through the activation of this signaling pathway. (C) Therapeutic effects of dsRNA mimicking miR‐152 (dsmiR‐152) or control non‐specific miRNA (dsNC) on tumor growth in vivo. Left panel: a schema indicating the protocol of in vivo analysis (upper) and bar graph showing effects of dsmiR‐152 on tumor growth in three SCID mice (lower). Right panel: photograph shows tumor‐bearing SCID mice (upper) and their subcutaneous tumors (lower) at the end of in vivo analysis. These findings strongly support the great potential of dsRNA mimicking miR‐152 to be applied to miRNA replacement therapy for cancers.
