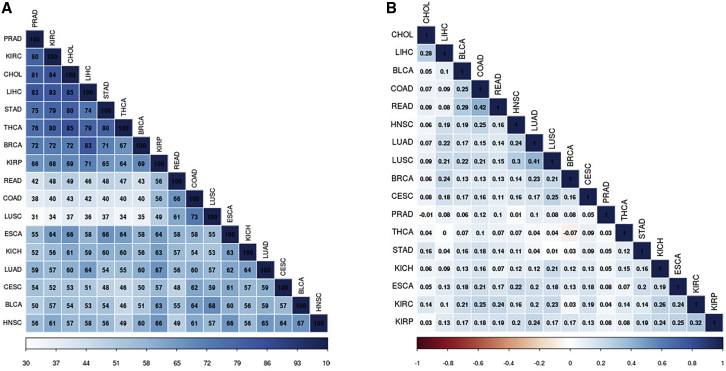
Figure 4.
Overlap Analysis between Cancer Cohorts
(A) Gene set enrichment analysis was performed on the top-25%-scoring kinases for each cancer cohort using MSigDB. Pairwise overlap of 100 gene sets was calculated. Cohorts that share the highest degree of gene set enrichments include LIHC and CHOL, KIRC and CHOL, and THCA and CHOL.
(B) Spearman-rank correlation between CKIs of each cancer cohort. The greatest degree of correlation was present between cancers of the same tissue, including COAD and READ or LUAD and LUSC. THCA and BRCA had a slight negative correlation, suggestive of opposing clinical relevance of the same kinases in the different cancer cohorts.