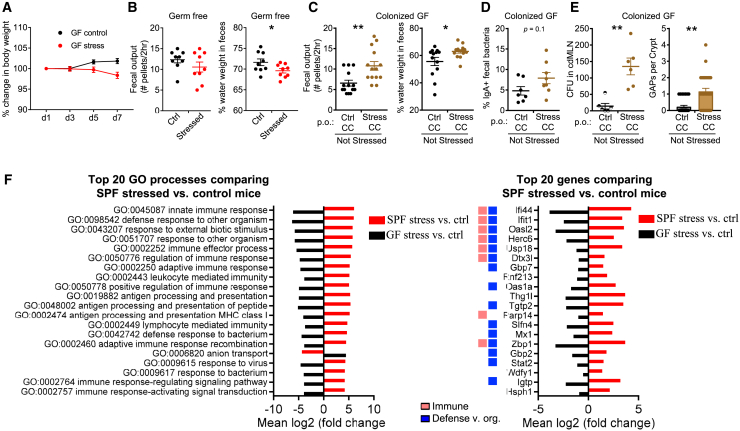
Figure 3.
Dysbiosis Induced by Stress Drives Intestinal and Immunological Alterations
(A and B) Effect of stress on germ-free (GF) mice. Stressed GF mice were assessed for the following: (A) change in body weight (n = 5, 1 expt.) and (B) GI function assessed by the number of fecal pellets expelled in a 2-h duration and percentage of fecal water weight on day 8 (n = 10, 2 expts.).
(C–E) Microbiota are sufficient to induce features of stress. GF mice were transplanted via oral gavage with stressed or control CCs without concurrent stress and assessed 7 days later for the following: (C) number of fecal pellets expelled in a 2-h duration and percentage of fecal water weight (n = 13, 2 expts); (D) percentage of IgA-bound fecal bacteria (n = 7–8, 2 expts); and (E) number of GAPs per crypt in the proximal colon and CFUs in cdMLN (n = 8–13, 1 expt).
(F) Stress-induced biological processes in the colon. GF and SPF mice were stressed for 7 days. The following day, the entire colon and cecum with respective unstressed controls was analyzed by RNA-seq (n = 3/group, 1 expt). The top 20 Gene Ontology processes created with Pathview with GAGE padj < 0.05 comparing SPF stressed versus control mice were chosen for display in red. The fold change for the same pathways (padj < 0.05) in GF stressed versus control mice is displayed for comparison (black). The top 20 most significant genes (padj < 0.05) that were upregulated or downregulated in SPF stressed versus control mice, with colors indicating the Gene Ontology processes categories in legend. All of the pathways with the category designations and involved genes can be found in Data S2.
The data are presented as means ± SEMs, Student’s t test, and 2–3 independent experiments for all of the panels unless otherwise specified. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0005. See also Figures S2 and S3.