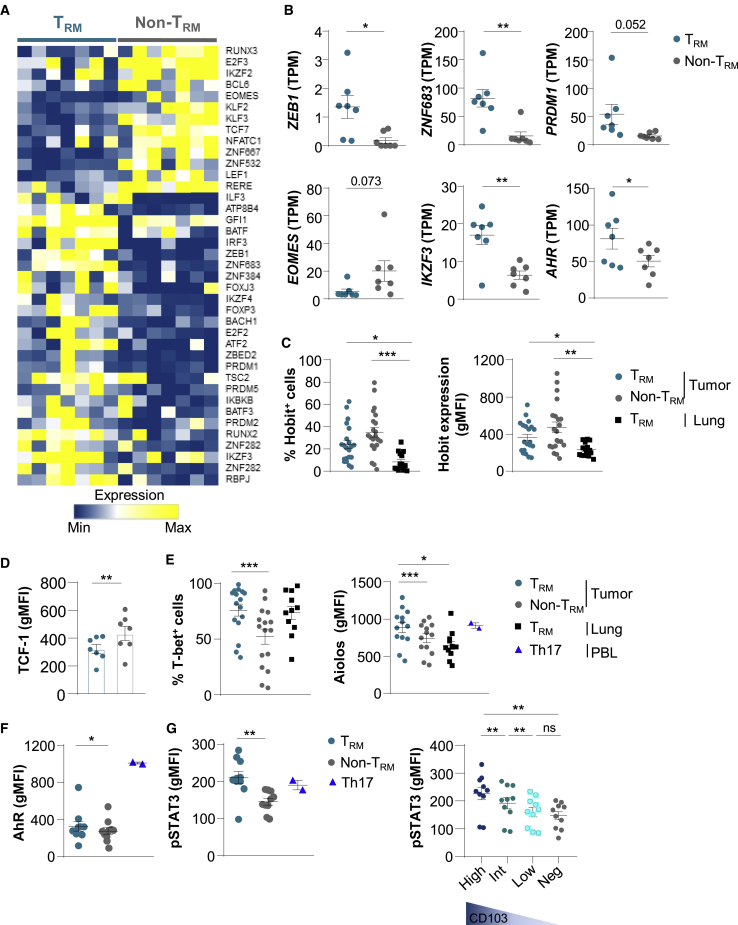
Figure 4.
Transcription Factor Profiles of TRM and non-TRM from NSCLCs
(A) Heatmap of transcripts encoding transcription factors differentially expressed in CD103+ and KLRG1+ CD8+ TIL (n = 7) (p < 0.05).
(B) Expression of ZEB1, ZNF683 (HOBIT), PRDM1 (BLIMP1), IKZF3 (AIOLOS), EOMES, and AHR genes in TRM and non-TRM from NSCLCs (n = 7). Values are transcripts per milion (TPM).
(C) Percentages of Hobit+ cells among TRM and non-TRM cells from NSCLCs (n = 21) and among healthy lung TRM cells (n = 15). Right, expression of Hobit (gMFI) in TRM and non-TRM cells from NSCLCs (n = 21) and in healthy lung TRM cells (n = 15).
(D) Expression of TCF-1 (gMFI) in tumor TRM and non-TRM cells (n = 7).
(E) Percentages of T-bet+ cells among tumor TRM and non-TRM cells (n = 21) and among healthy lung TRM cells (n = 11). Right, expression of Aiolos (gMFI) among tumor TRM and non-TRM and paired healthy lung TRM (n = 13). Heathy donor Th17 control cells are included (n = 2).
(F) Expression of AhR (gMFI) in tumor TRM and non-TRM (n = 10), and in Th17+ control cells (n = 2).
(G) Expression of pSTAT3 (gMFI) in tumor TRM and non-TRM cells (n = 10), and in Th17+ controls (n = 2). Right, expression of pSTAT3 (gMFI) in tumor TRM cells displaying CD103high (High), CD103int (Int), and CD103low (Low) phenotype, and in CD103−CD8+ TILs (Neg) (n = 10).
Each symbol represents an individual donor; horizontal lines are means ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (paired t test or ANOVA with Bonferroni post hoc test). gMFI, geometric mean fluorescence intensity; ns: not significant.