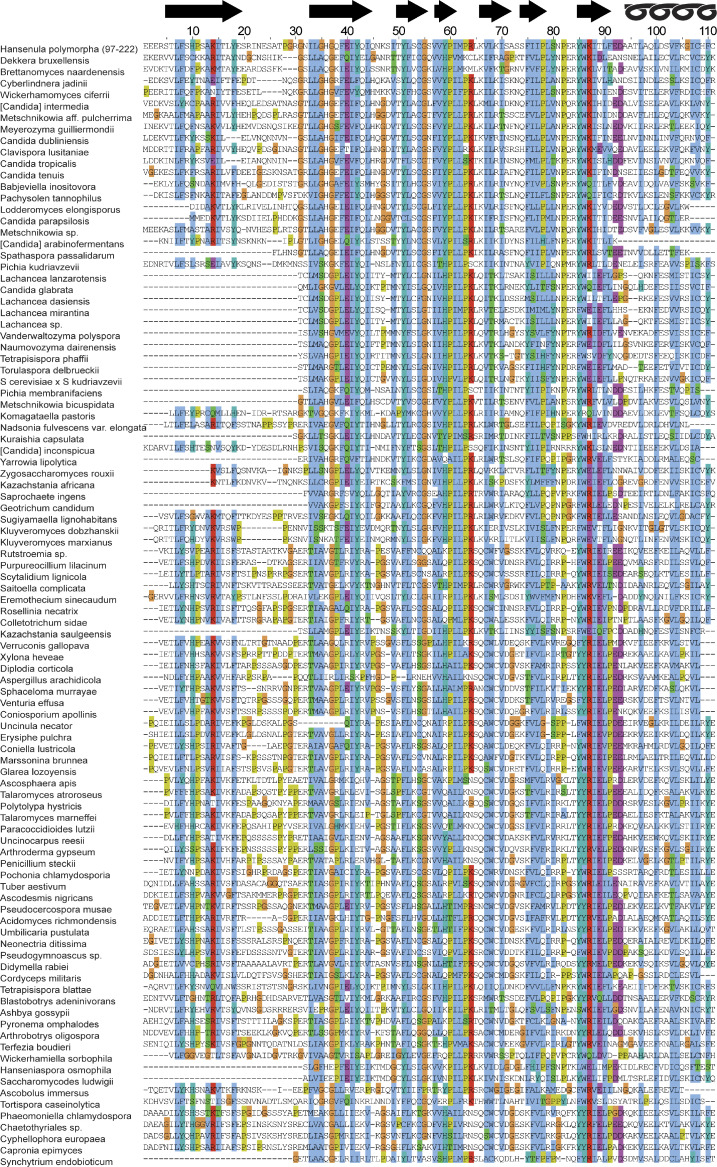
Figure S1.
The multiple sequence alignment obtained by HHblits with standard settings (three iterations, searching into a UNiREF database at 30% nonredundancy) contained Inp1 from H. polymorpha (residues 97–222) and 97 other sequences. These were aligned to remove all gaps in the seed sequence, and colored according to the CLUSTAL scheme, with predicted strands 1 to 7 and α helix indicated at the top. Note the lack of conserved positively charged residues from any loop.