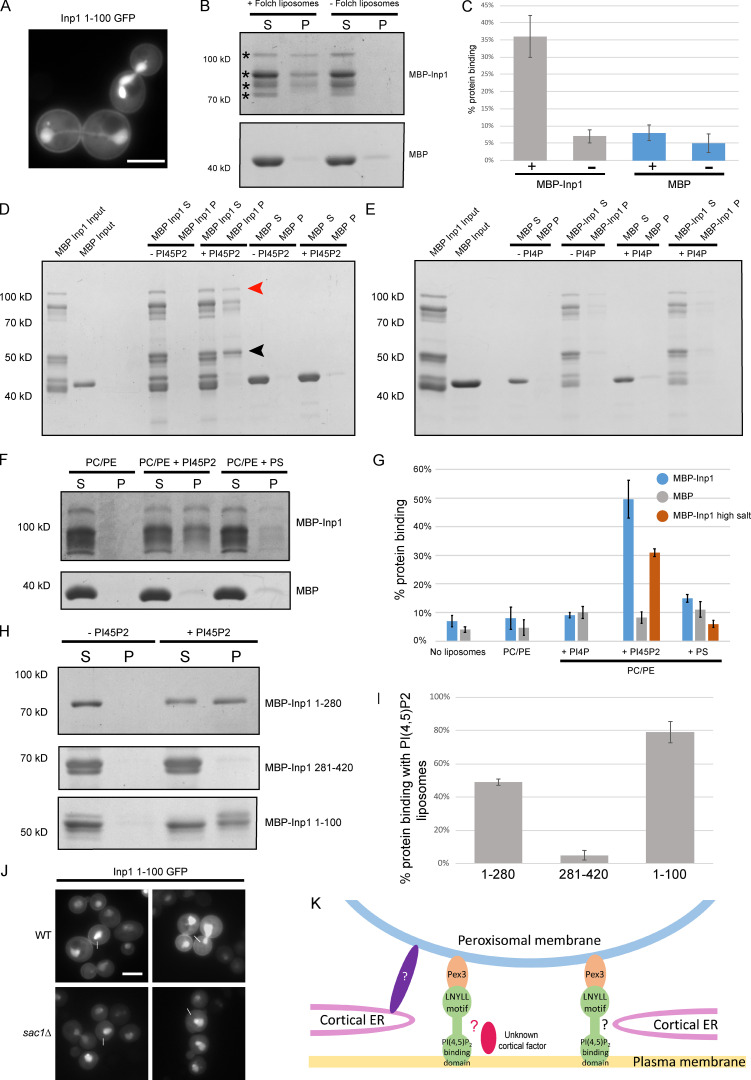
Figure 5.
Inp1 has an N-terminal PI(4,5)P2 binding domain. (A) An inducible truncation of 1–100 Inp1-GFP was expressed under control of the GAL1/10 promoter in WT cells and examined by epifluorescence microscopy. A single focal plane is shown. (B) Purified MBP-Inp1 was incubated with Folch fraction I liposomes and subjected to ultracentrifugation. Supernatant (S) and pellet (P) fractions were then analyzed by SDS-PAGE and Coomassie staining. Asterisks indicate multiple MBP-Inp1 fragments. (C, G, and I) Each supernatant and pellet band when combined were considered to represent 100% of the protein used in the experiment. + and − indicate the presence or absence of liposomes, respectively. For MBP-Inp1 high salt, purified MBP-Inp1 was incubated with liposomes made up of PC, PE with or without either PI(4,5)P2 or PS in the presence of 370 mM salt (20 mM KCl + 350 mM NaCl). Liposome-binding assays were all performed in at least three independent experiments. Error bars represent SEM. (D–F) Purified MBP-Inp1 or MBP only were incubated with liposomes made up of PC, PE with or without 22% PI(4,5)P2 (D and F), PI(4)P (E), or PS (F) and subjected to ultracentrifugation. Total protein input, supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and Coomassie staining. Red arrowhead indicates full-length MBP-Inp1. Black arrowhead indicates a breakdown product truncation of MBP-Inp1 that is still able to bind to lipids containing PI(4,5)P2. (H) Purified truncations of MBP-Inp1 were incubated with liposomes made up of PC, PE with or without 22% PI(4,5)P2 and subjected to ultracentrifugation. Supernatant (S) and pellet (P) fractions were then analyzed by SDS-PAGE and Coomassie staining. (J) An inducible truncation of 1–100 Inp1-GFP was expressed in WT and sac1Δ cells and examined by epifluorescence microscopy. White lines indicate where measurements for line-scan analyses were taken from (see Fig. S3 G). Single focal planes are shown. Multiple images from the same experiment are shown. Scale bar, 5 µm. (K) Models of peroxisome tethering to the mother cell cortex. Inp1 is a PM-PER tether that bridges peroxisomes and the plasma membrane by interacting with peroxisomal Pex3 via a conserved LNYLL motif and with the plasma membrane via an N-terminal domain that binds PI(4,5)P2. This PM-PER tether is required for peroxisome retention. Additional peroxisome contacts with cortical structures may contribute to peroxisome positioning or distribution at the mother cell cortex. Interaction with the cER could occur via Inp1 and not-yet-identified factors (indicated by black question mark). Inp1 could also be in contact with cortical structures/factors independent of the cER (indicated by the red oval and red question mark). Peroxisome contact with the cER could also occur independently of Inp1 and via additional unidentified factors (indicated by the purple oval and white question mark).