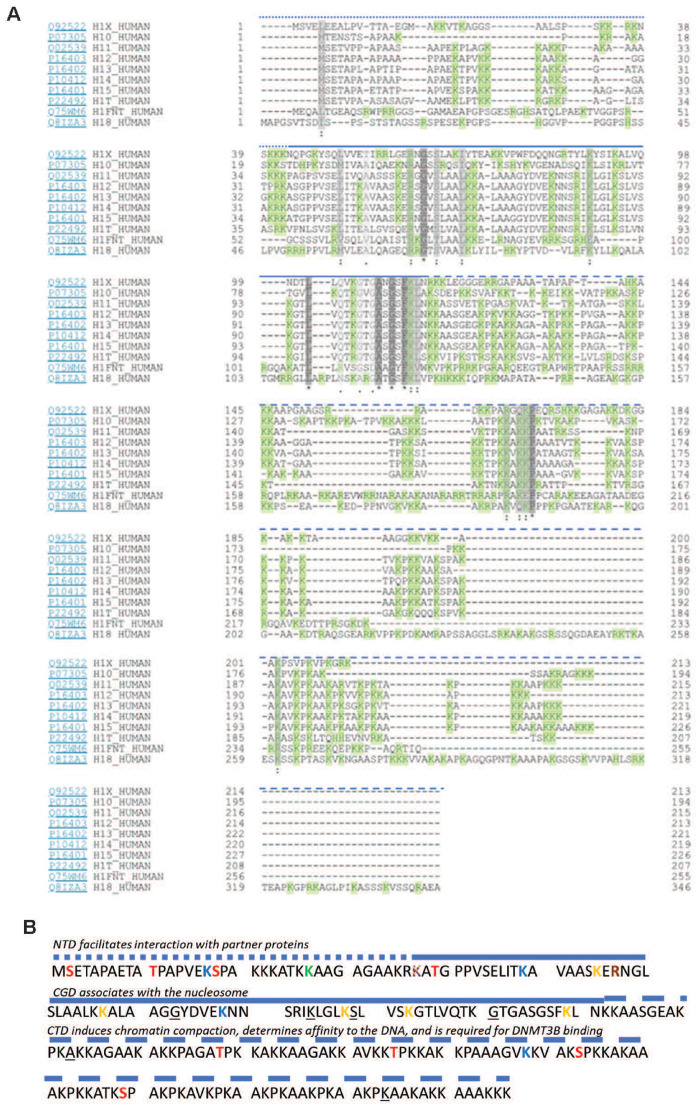
Figure 1:
Comparison of human histone 1 isoforms with a flashlight on H1.5. (A) Alignment of the 11 linker histone subtypes. Grey—similarity (the darker, the greater the extend of similarity between subtypes). Green—positively charged amino acid residues. Heavily dashed blue line, N-terminus; straight blue line, globular domain; lightly dashed blue line, C-terminus [based on Uniprot and NCBI protein-tools as well as (1)]. (B) H1.5 protein sequence. Phosphorylated residues (marked red): T10, S17, T39, T138, T155, S173, S189. Methylated residues (marked green): K27. Acetylated residues (marked blue): K17, K49, K78, 168. β-Hydroxybutyrylated residues (marked yellow): K37, K55, K67, K88, K93, K109. Succinylated residues (marked purple): K37. Citrullinated residues (marked orange): R57. Sites mutated in lymphoma patients are underlined: 73G, 84K, 89S, 101G, 123A, 214K. NTD, N-terminal domain; CGD, central globular domain; CTD, C-terminal domain. N-terminus, straight blue line; globular domain, lightly dashed blue line; C-terminus [based on Uniprot and NCBI protein-tools as well as (1, 29)]