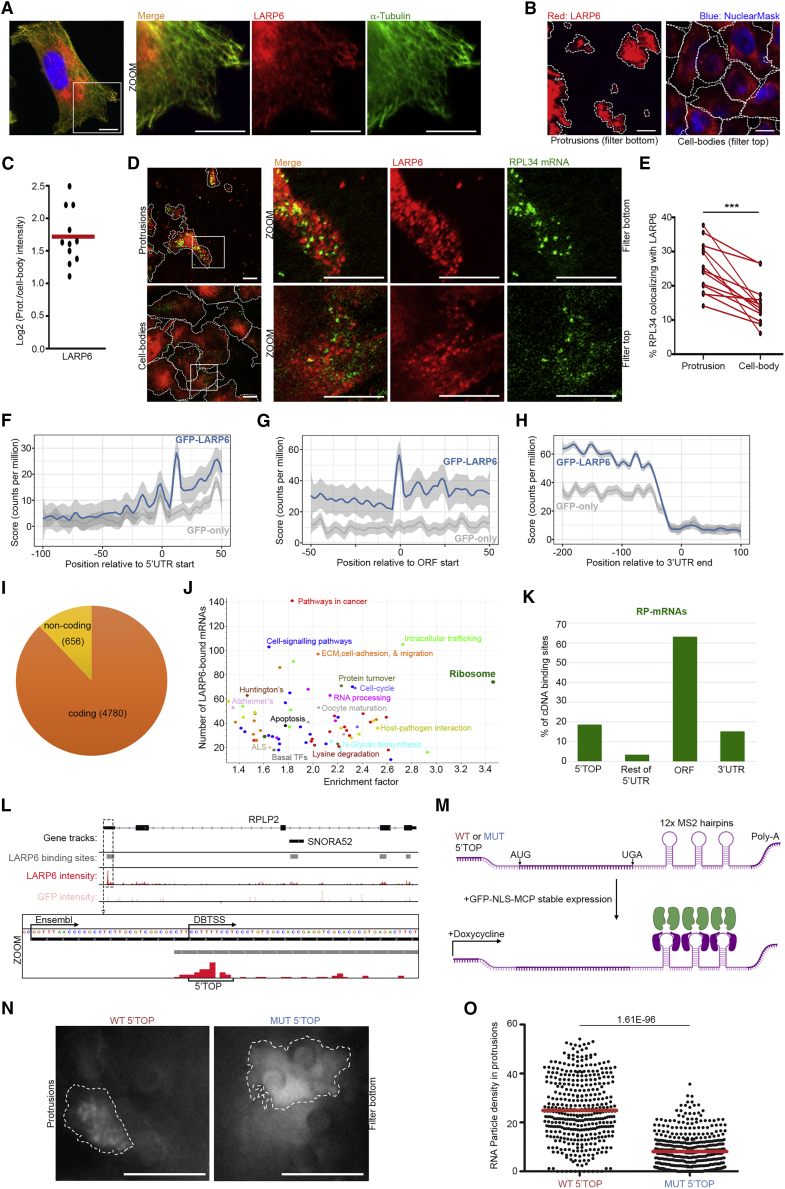
Figure 3.
Transcriptome-wide iCLIP Studies Reveal Direct Binding of LARP6 to RP-mRNAs
(A) LARP6 is localized to cytoplasmic puncta that track microtubules. Representative IF images of LARP6 (red) and α-tubulin (green) in MDA-MB231 cells grown on collagen-coated slides. Nucleus was stained with NuclearMask (blue).
(B) LARP6 puncta are enriched in protrusions. Representative IF images of LARP6 (red) in protrusions and cell bodies of MDA-MB231 cells. Cell boundaries (dashed lines) were defined from co-staining with anti-tubulin antibody.
(C) Quantification of IF images from experiments shown in (B), revealing LARP6 enrichment in protrusions. A total of 11 large field of view images, measured from 2 independent experiments were quantified.
(D) LARP6 co-localizes with RP-mRNAs in protrusions. Representative RNA-FISH and IF co-staining images of RPL34 mRNA (green) and LARP6 (red) in protrusions and cell bodies of MDA-MB231 cells. Cell boundaries (dashed lines) were defined from co-staining with anti-tubulin antibody.
(E) Quantification of the % of co-localization of RPL34 mRNA with LARP6 in corresponding protrusion and cell-body images from experiments shown in (D). A total of 13 large field of view images from 2 independent experiments were quantified. Red lines connect values of protrusion and body from the corresponding images. p values were calculated using a two-tailed, homoscedastic t test. ∗∗∗p < 0.001.
(F) Metaprofile plot of LARP6 iCLIP crosslink sites at the aligned annotated intergenic-5′UTR junctions (2,204 landmarks), showing preferential association with specific regions at the vicinity of TSS.
(G) Metaprofile plot of LARP6 iCLIP crosslink sites at the aligned annotated 5′UTR-ORF junctions (4,122 landmarks), showing preferential association with the translation start site.
(H) Metaprofile plot of LARP6 iCLIP crosslink sites at the aligned annotated 3′UTR-intergenic junctions (6,333 landmarks), showing association throughout the 3′UTR.
(I) LARP6 mainly binds protein-coding transcripts. Pie chart showing the prevalence of coding versus non-coding RNAs among LARP6 binding targets (Dataset S6).
(J) The KEGG category of ribosome (green), which is comprised all RP-mRNAs, is significantly enriched among LARP6-binding targets. Fisher’s exact test analysis (FDR < 0.02) of mRNA categories, which are significantly over-represented among the identified LARP6 targets. Each data point represents a functional category from KEGG database, with similar categories highlighted by the same colors (Dataset S7).
(K) LARP6 interacts with RP-mRNAs via multiple regions. Distribution of LARP6-binding regions in RP-mRNAs.
(L) An example genomic view of LARP6-specific binding sites after peak calling (gray tracks) in an RP-mRNA (RPLP2), along with read intensities for GFP and GFP-LARP6 iCLIP runs. Four distinct LARP6-binding sites are mapped to the RPLP2 locus: two mapping to the ORF region, one to RPLP2 3rd intron, which is annotated as SNORA52, and one to the 5′UTR. Inset: zoomed view of RPLP2 5′UTR showing the LARP6-binding site overlapping with the 5′TOP. Note that for most RP-mRNAs, annotation of TSS in Ensembl is further upstream of the more accurately annotated DBTSS (Suzuki et al., 2018).
(M) Schematic representation of the MS2 reporter system for live-cell monitoring of 5′TOP mediated RNA localizations.
(N) WT 5′TOP motif is sufficient for RP-mRNA localization to protrusions. Representative still images of the GFP-MCP signal in transwell protrusions of WT or MUT 5′TOP reporter engineered MDA-MB231 cells described in (M), following induction of reporter expression with 2 μg/mL doxycycline for 12 h. GFP-MCP exhibits a punctate pattern in protrusions of WT 5′TOP reporter expressing cells, indicative of association with mRNA particles, as opposed to a diffuse pattern in protrusion of MUT 5′TOP reporter expressing cells.
(O) Quantification of mRNA particles in protrusions of WT-5’TOP versus MUT-5’TOP reporter expressing cells from experiments shown in (N). A total of 25 (WT) and 28 (MUT) time-lapse videos (3 s at 0.2-s intervals) from 2 independent experiments were quantified. The number of discrete particles identified at every frame image were quantified and normalized to the protrusion area to determine mRNA molecule density. The p value was calculated using a two-tailed, homoscedastic t test. All scale bars, 10 μm.