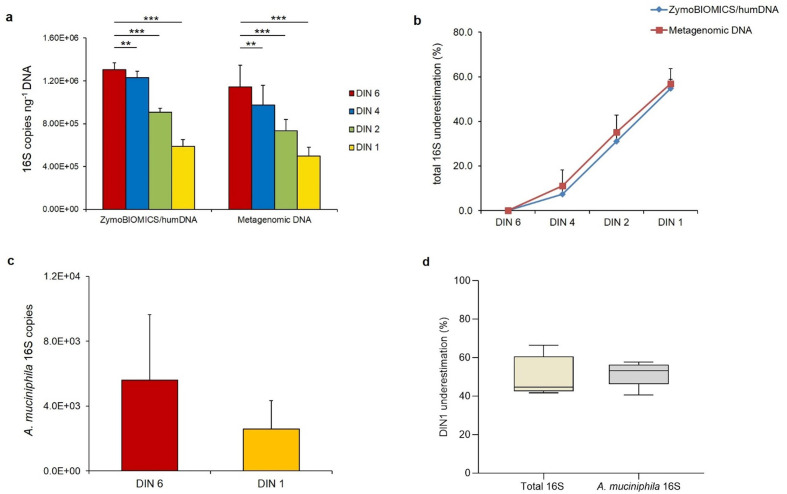
Fig. 3.
Total and species-specific 16S rRNA gene copy number quantification is affected by metagenomic DNA quality. (a) 16S copy quantification in 13 metagenomic DNAs at their original DIN6 value and at three degradation levels, corresponding to DIN4, DIN2 and DIN1 values, compared to the ZymoBIOMICS/humDNA mock at the same integrity levels. (b) 16S copies underestimation percentage for the ZymoBIOMICS/humDNA mock and for the 13 metagenomic DNAs at the same integrity levels. (c) A. muciniphila 16S copies measured in five metagenomic DNAs (f9-f13) at their original DIN6 value and at the DIN1 degradation level. (d) A. muciniphila 16S copy underestimation percentage (56%) is in accordance with that measured for total 16S copies in the same metagenomic DNAs (f9–f13) at the DIN1 degradation level. 16S copies are reported as the mean of at least two replicates for each sample±sd. 16S copy underestimation was calculated as a percentage compared to the highest quality (DIN 6) samples (considered as 100%). *, P<0.05, **,P<0.01, ***, P<0,001 vs DIN6.