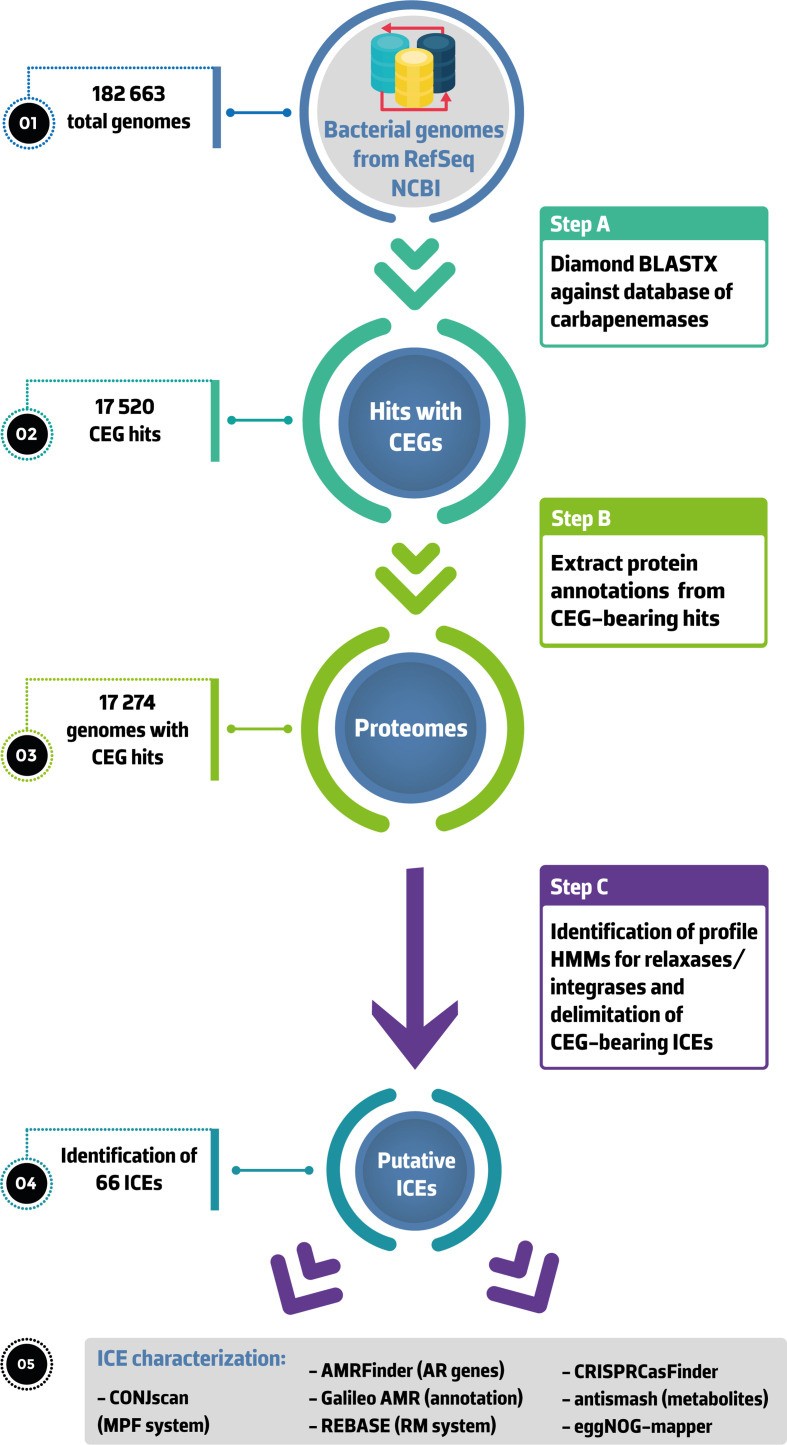
Fig. 1.
Overview of the workflow followed in this study. All assemblies available in NCBI RefSeq were downloaded and blasted against an in-house database of carbapenemases using diamond blastx (step 1). NCBI annotated proteins from CGE-bearing genomes were then extracted (step 2) and used for the identification of relaxase and serine or tyrosine recombinase (step 3). Search of directed repeats and delimitation of putative ICEs was also performed. CONJscan was used to identify the MPF family of each element. We then looked for AR genes, restriction–modification systems, CRISPR arrays and their associated (Cas) proteins, as well as secondary metabolites within extracted ICEs. We also characterized the functional annotations of their proteomes and the MLST of the genomes carrying a CEG-bearing ICE. Abbreviations: AR, antibiotic resistance; CEG, carbapenemase-encoding gene; HMM, hidden Markov model; ICE, integrative and conjugative element; MPF, mating-pair formation; RM, restriction–modification.