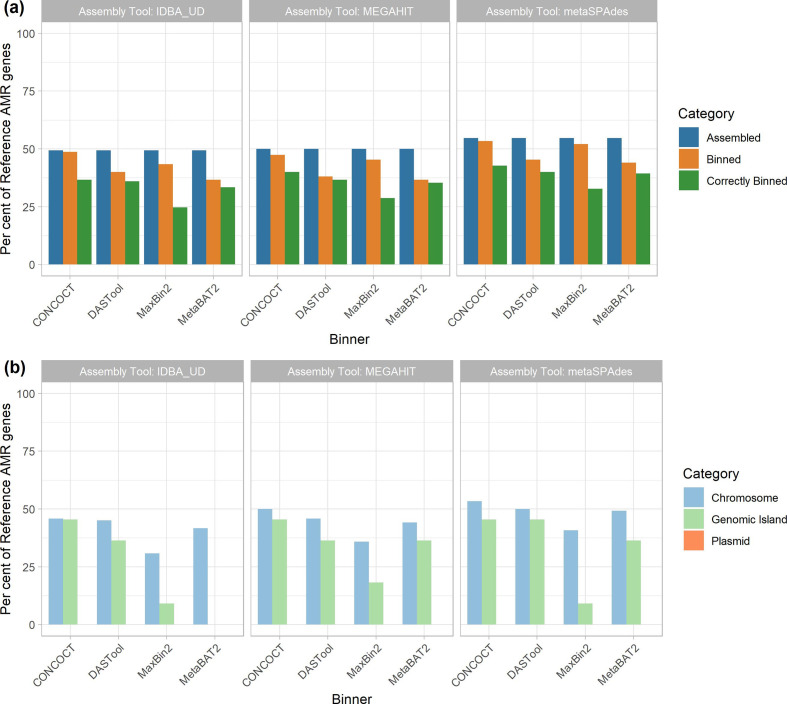
Fig. 5.
Recovery of AMR genes across assemblers, binners and genomic context. (a) The proportion of reference AMR genes recovered (y-axis) was largely similar across assembly tools (panels as indicated by title) at roughly 50 %, with metaSPAdes performing marginally better overall. Binning tools (x-axis) resulted in a small reduction in AMR genes recovered (orange); however, only 24–40 % of all AMR genes were correctly binned (green). metaSPAdes- CONCOCT was the best performing MAG binning pipeline. (b) Per cent of correctly binned AMR genes recovered by genomic context. MAG methods were best at recovering chromosomally located AMR genes (light blue) regardless of metagenomic assembler or binning tool used. Recovery of AMR genes in GIs showed a bigger variation between tools (light green). None of the 12 evaluated MAG recovery methods were able to recover plasmid-located AMR genes.