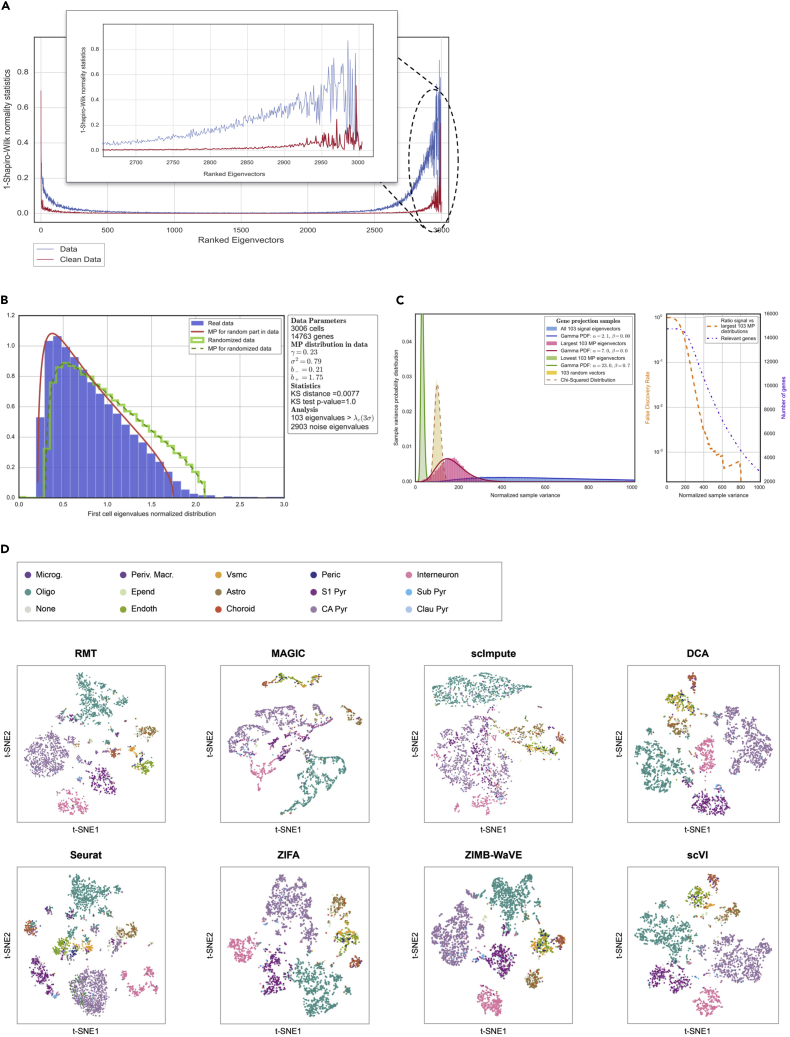
Figure 5.
Application to Mouse Cortex Single-Cell Expression
(A) Localization properties of the eigenvectors in a single-cell dataset of PBMCs.32 The blue line represents the system dominated by sparsity and the red line corresponds to the system after removing sparsity. This figure also shows how some eigenvectors corresponding to eigenvalues out of MP distribution are delocalized (red line) and therefore do not carry any information.
(B) MP prediction and identification of relevant components.
(C) Study of the chi-squared test for the variance (normalized sample variance) in signal and noise gene projections. In the left panel, the distributions correspond to a projection of genes into the 103 signal eigenvectors (corresponding to the 103 eigenvalues of A) and the projection into the 103 lowest and 103 largest MP eigenvectors. There is also a projection into 103 random vectors. Finally, the lines show how gamma functions can fit the distributions discussed. The right panel shows the number of relevant genes in terms of the test discussed above together with a false discovery rate. Higher values for the chi-squared test for variance indicate that the genes are less responsible for the signal.
(D) Comparison of the t-SNE representation for different methods and algorithms. This case corresponds to 15 different mouse cortex cell phenotypes described in Zeisel et al.34