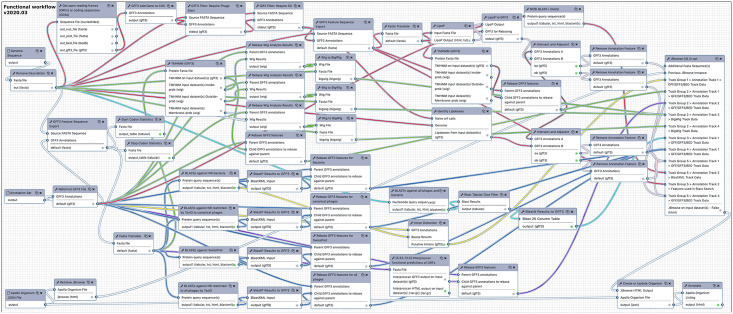
Fig 3. The functional workflow links tools in Galaxy used for functional prediction.
Inputs for the functional workflow are gene calls paired with their genome. These are piped through five sub-paths within the analysis. 1) The BLASTn path uses full genomic nucleotide sequence (light blue noodles). 2) The BLASTp protein analysis against curated (UniProtKB SwissProt) and sequence-inclusive databases (NCBI nonredundant (nr)) (dark blue noodles). 3) The search for interrupted genes like introns compiles separate CDS hits to the same protein (yellow noodles). 4) A directed search for spanin proteins using TMHMM, lipobox-finding (using LipoP and a motif search), and BLASTp against a curated database (magenta and pink noodles). 5) Domain analysis plots comprehensive TMHMM outputs and InterProScan results for conserved domains and signatures (green and purple noodles, respectively).