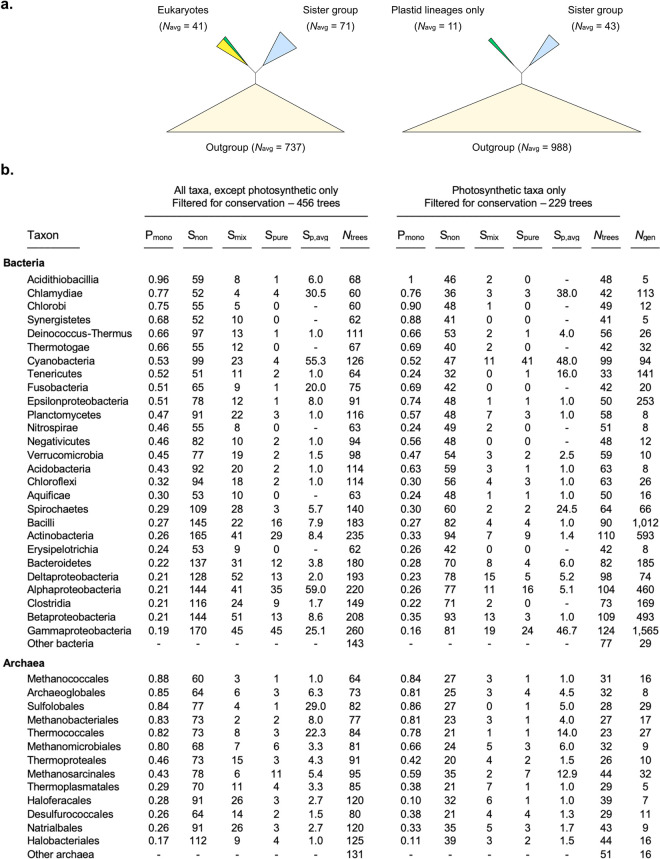
Fig 4. Identification of the prokaryotic sister group to the eukaryotes in 2,575 eukaryotic-prokaryotic unrooted gene trees (EPC).
a. shows the average clade sizes for eukaryotes, the sister group to eukaryotes and the outgroup in the analyzed trees for (right) the 229 trees with only plastid derived lineages and (left) for the 456 EPCs containing all taxa except photosynthetic lineages. b. details the list of bacterial (top) and archaeal (bottom) phyla occurring in the trees with only plant lineages (right) and all other trees (left) that were filtered for conservation (average branch length of the tree ≤ 0.1). Archaeal and bacterial phyla with less than 5 representative species in the dataset were collapsed into ‘other archaea’ and ‘other bacteria’ groups. Pmono refers to the proportion of trees with a branch (split) separating the species of the respective phylum from all the others in the tree; Snon refers to the number of occurrence of the phylum only in the outgroup clade; Smix refers to the number of occurrences of the phylum as a mixed sister (more than one phylum in the clade); Spure refers to the number of occurrences of the phylum as pure sister (as the single phylum); Sp,avg shows the average size of the sister clade when the phylum occurs as a pure sister clade. Ntrees show the number of occurrences of the phyla across the trees and Ngen indicates the number of species in each taxon included in the complete dataset.