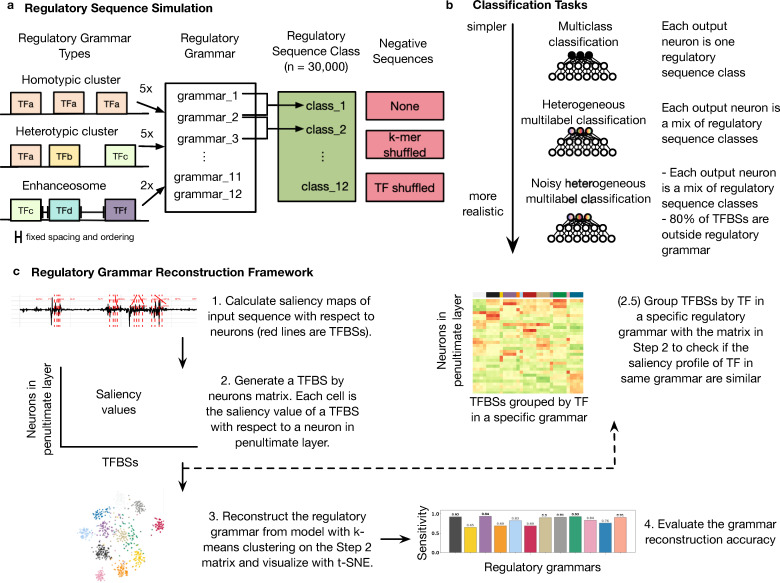
Fig 1. Pipeline for analyzing regulatory grammar learned by ResNet models trained on simulated regulatory sequences.
(a) Regulatory sequence and negative sequence simulation. We designed twelve regulatory grammars, including five homotypic clusters, five heterotypic clusters, and two enhanceosomes as prototypes for simulated regulatory sequences. Then, to reflect that regulatory regions active in a cellular context may have multiple grammars, we defined twelve regulatory sequence classes, each with two different grammars. Finally, we generated two sets of negative sequences: k-mer shuffled and TF shuffled versions of the simulated positive sequences. (b) Classification tasks. ResNets are trained on simulated regulatory sequences and the negative sets in three increasingly realistic scenarios. (c) Regulatory grammar reconstruction framework.