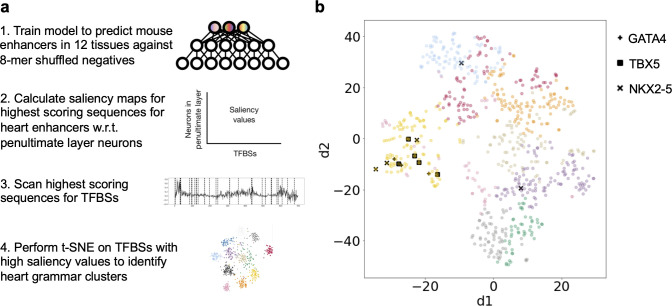
Fig 7. ResNets identify known heart heterotypic cluster when trained on mouse enhancers.
We trained a ResNet on developmental mouse enhancers from 12 tissues identified from histone modifications (S6 Fig) and applied our saliency map approach to interpret the trained network. a) Pipeline for identifying regulatory grammar in mouse developmental heart enhancers. b) t-SNE visualization of clustered TFBS saliency maps from top scoring heart enhancer sequences. Clusters determined by k-means with k = 9 are indicated by color (S6C Fig). Instances of NKX2-5, TBX5, and GATA4 motifs are labelled with shapes. These factors form an essential heterotypic cluster during heart development and are significantly enriched in cluster 4 (S5 Table).