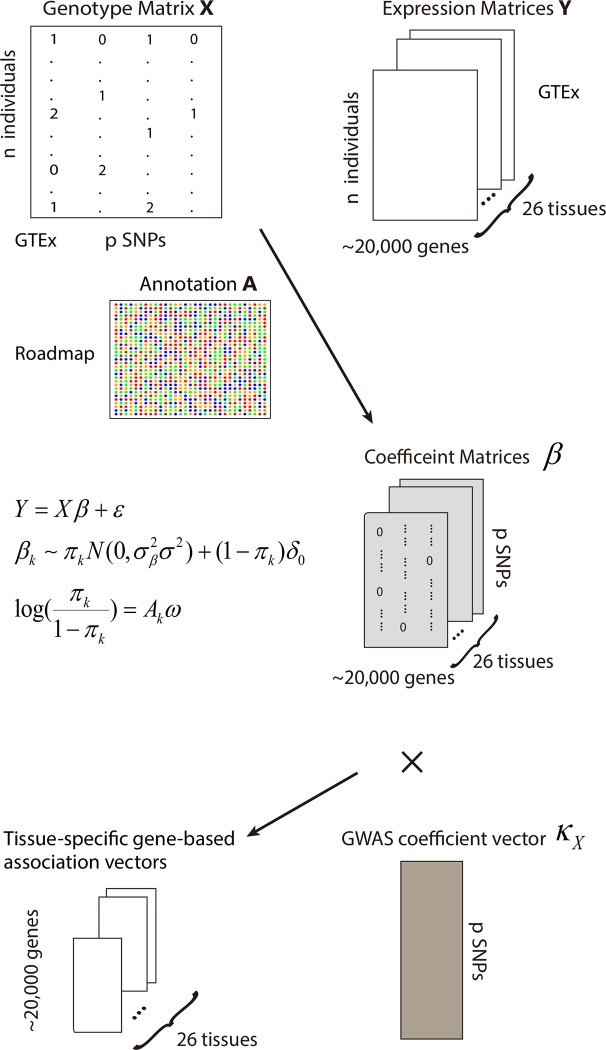
Fig 1. The general scheme of our method.
Gene expression imputation models were built based on gene expression matrix Y of a specific gene in a tissue, the genotype matrix X and epigenetic annotation matrix A of cis-SNPs of the gene. The annotation matrix was used to select SNPs having regulatory effects on gene expression since we assumed that only part of cis-SNPs have effects on gene expression of the nearby gene. After getting SNP coefficient vectors β in each tissue for the gene, we combined β’s with GWAS summary stats and then get the gene-level association statistics for each disease in each tissue.