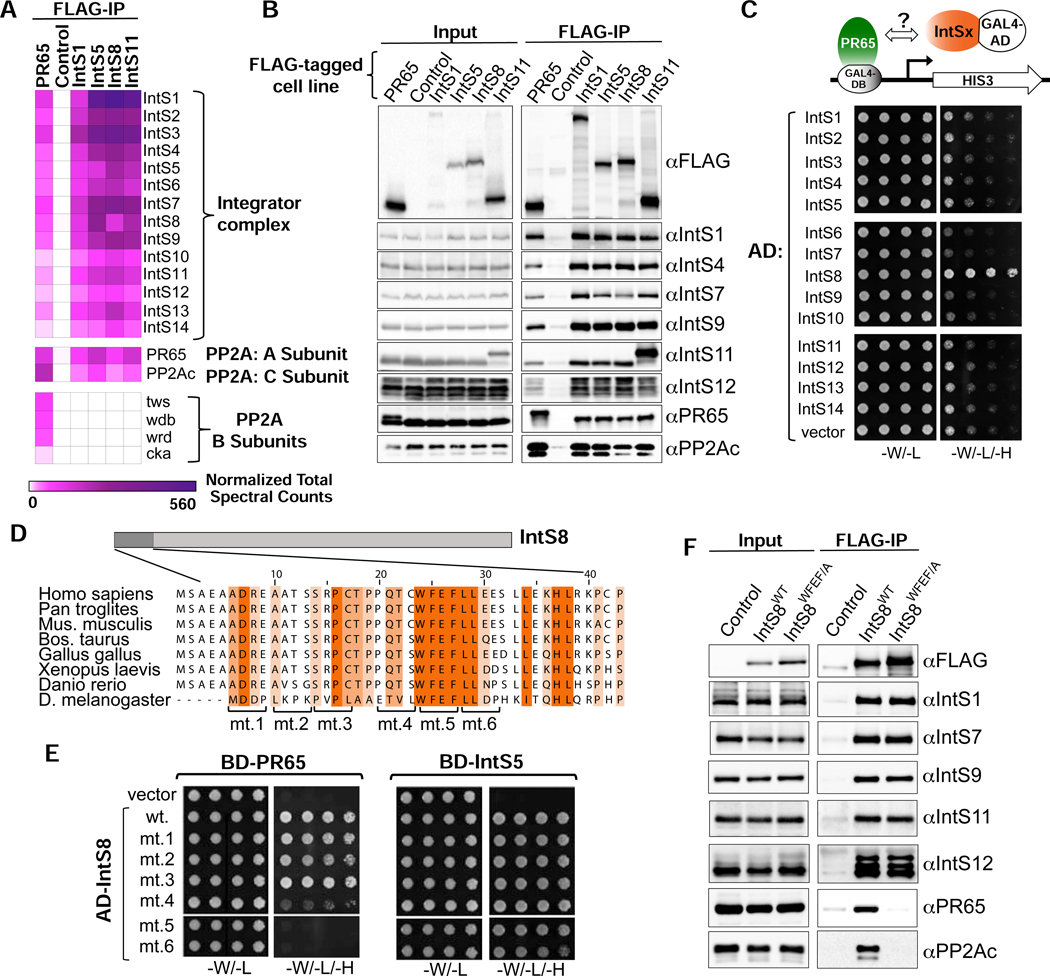
Figure 2. PR65/PP2Ac require a discrete motif within IntS8 to associate with Integrator.
(A) Heatmap derived from IP LC-MS analysis of FLAG immunoprecipitations from S2 cells expressing the indicated FLAG-tagged proteins. Heatmaps reflect normalized spectral counts observed from analysis of samples performed in triplicate. Control cells lack any exogenously expressed FLAG-tagged protein.
(B) Western blot analysis of input nuclear extracts (left) and immunoprecipitations (right) from S2 cells stably expressing FLAG-tagged proteins as indicated. Immunoprecipitations conducted using anti-FLAG affinity resin were normalized to FLAG signals in each immunoprecipitation.
(C) Yeast two-hybrid analysis where PR65 was fused to the Gal4 DNA binding domain and 14 Integrator subunits were individually expressed as fusions with the Gal4 activation domain. Empty vector is the negative control. Permissive plates lack leucine and tryptophan (-L/-W) to allow growth of plasmid-containing yeast whereas selective plates also lack histidine to screen for interaction (-L/-W/-H). Yeast are plated in five-fold serial dilutions.
(D) Alignment of the N-terminus of IntS8 from multiple species. Scanning mutations are depicted where four consecutive amino acids are converted to alanine. Note the numbering is relative to Homo sapiens INTS8.
(E) Results of yeast two-hybrid analysis where either PR65 or IntS5 are fused to the Gal4 DNA binding domain and either wild-type or alanine mutant IntS8 constructs are fused to the Gal4 activation domain.
(F) Western blot analysis of nuclear extracts (left) and immunoprecipitations (right) from stable cell lines expressing FLAG-IntS8-WT or FLAG-IntS8-WFEF/A.