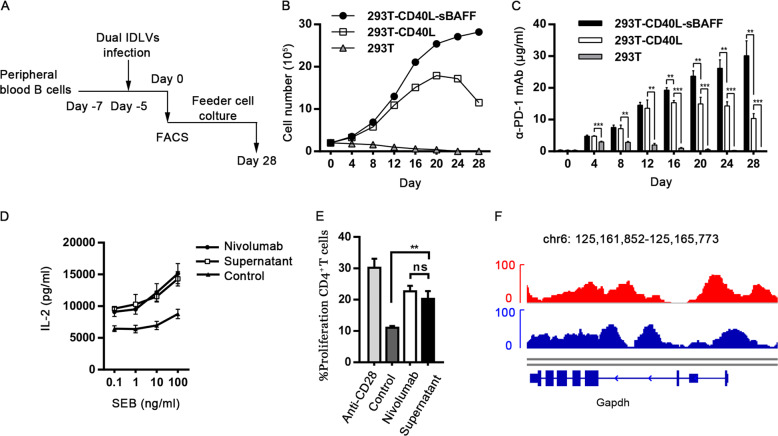
Fig. 3. Functional α-PD-1 mAb secretion from engineered CD90-expressing B cells.
a Schematic representation of sorting and expansion strategy of engineered CD90-expressing B cells. b Following FACS sorting at day 0 indicated in a, engineered CD90-expressing B cells were co-cultured with the irradiated 293T, 293T-CD40L, or 293T-CD40L-sBAFF feeder cells. The feeder cells were renewed every 4 days and the numbers of B cells were counted to indicate expansion patterns. c As performed in a, culture supernatants of gene-edited B cells co-cultured with feeder cells were collected at various time points, followed by ELISA for detecting α-PD-1 mAb concentration. Data are representative of three independent experiments. d PD-1 blockade-mediated T-cell stimulation assay was performed by SEB stimulation of PBMCs. 1 × 105 PBMCs were stimulated with serial dilutions of SEB in the presence of nivolumab, culture supernatant of gene-edited B cells or untransduced B cells as a control. Supernatants were collected 3 days later and measured for IL-2 levels by ELISA. Nivolumab was used as a positive control. Representative data from three healthy donors are shown. e To conduct a T-cell proliferation assay mediated by PD-1 blockade, PBMCs from healthy donors were stimulated with anti-CD3 antibody and cultured in the presence of anti-CD28, nivolumab, culture supernatant of gene-edited B cells or untransduced B cells as a control for 3 days. The CFSE labeled CD4+ T cells were detected via flow cytometry. f Genome alignment tracks of the normalized ATAC-seq data showed the open chromatin for GAPDH locus in CD90+ engineered B cells cultured at day 28 (red). Pre-sitmulated B cells with only the donor IDLV were used as the control (blue). The results in panels c, d, and e are presented as mean ± SEM, n = 3. **P < 0.01, ***P < 0.001, ns, no significant difference; one-way ANOVA with Tukey’s post hoc tests (c–e) were used.