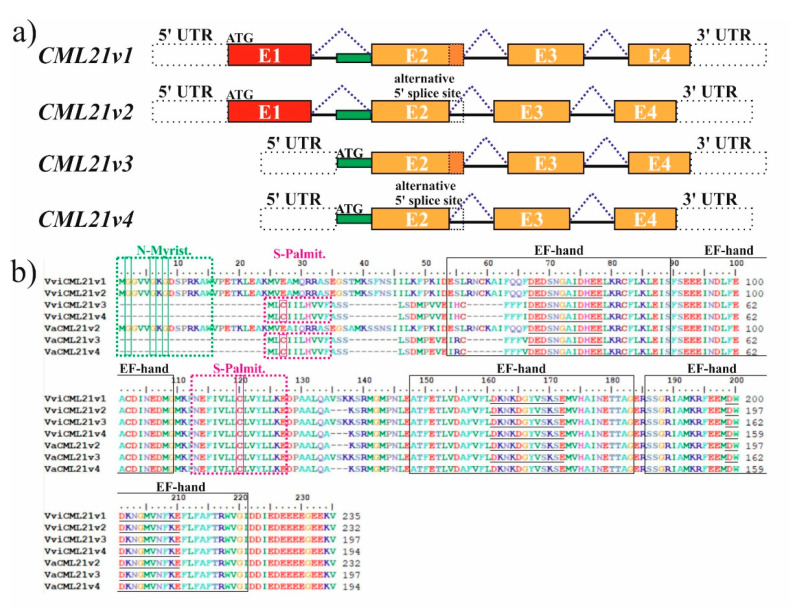
Figure 1.
The structure representation and protein sequence analysis of CML21 mRNA splice variants in grapevine. (a) Alternatively spliced CML21 transcript variants of Vitis amurensis identified in this work (VaCML21v2, VaCML21v3, VaCML21v4) and CML21 transcript variants of Vitis vinifera (VviCML21v1, VviCML21v2, VviCML21v3, VviCML21v4) predicted by genome sequence analysis and retrieved from the databases. Exons and introns are shown using boxes and lines, respectively, with white dashed boxes representing untranslated regions (UTRs). (b) Alignment and conserved motif analysis of CML21 deduced proteins in V. amurensis and V. vinifera. The EF hands were predicted by PROSITE scan [31,32] and the lipid modification sites were predicted using GPS-Lipid [33,34]. Multiple sequence alignment was conducted using BioEdit software [35].