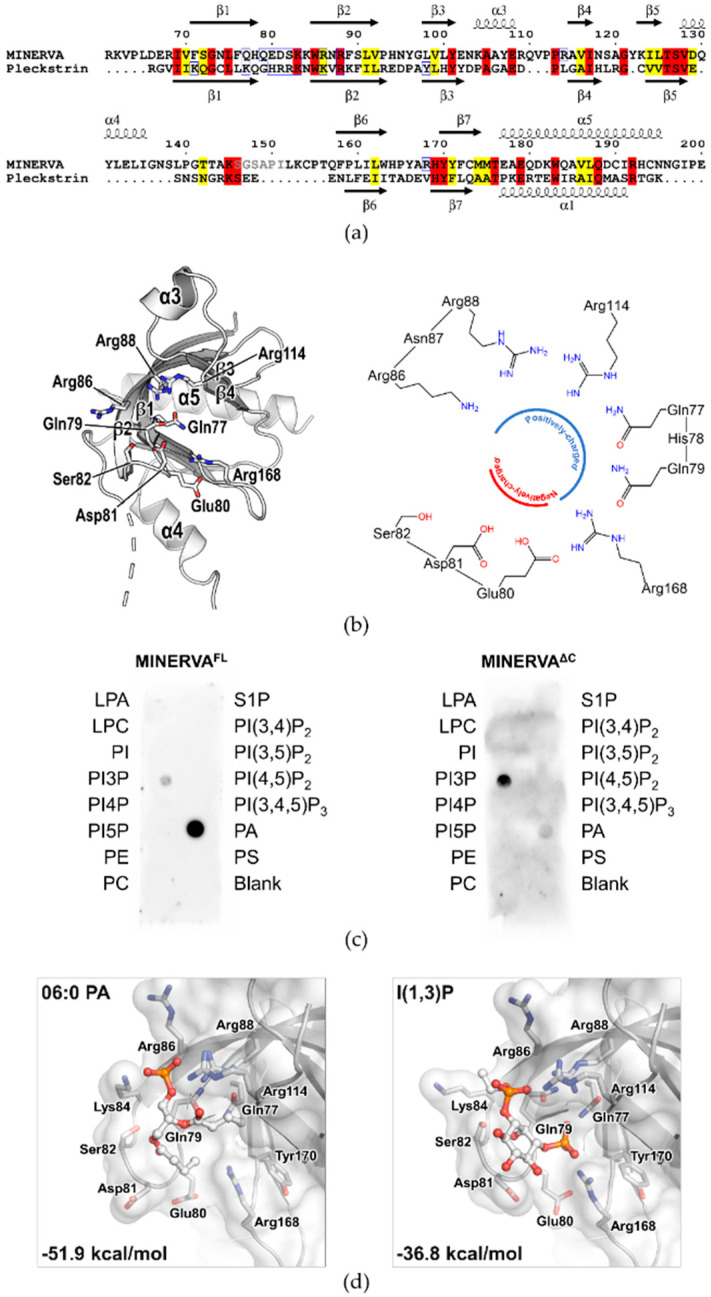
Figure 3.
The PH domain of MINERVA. (a) Structure-based sequence alignment of PH domains from MINERVA and Pleckstrin (PDB ID: 2I5F) showing conserved residues at the ligand binding site. Conserved residues and similar residues are highlighted in red and yellow, respectively. Residues contributing to ligand recognition are indicated with blue frames. Secondary structure elements of the MINERVA and Pleckstrin models are shown above and below the alignment, respectively. Unmodeled residues (Ser146–Ile151) of MINERVA due to lack of electron density around the cleavage site are in gray. (b) Left, cartoon representation of the PH domain of MINERVA with side chains of putative ligand interacting amino acids shown in stick representation. There are both positively charged and negatively charged residues in the ligand binding pocket. Right, schematic of the residues at ligand binding pocket of MINERVA PH domain showing positively charged and negatively charged faces. (c) Lipid dot-blot assay of MINERVAFL (left) and MINERVAΔC (right). Both MINERVALFL and MINERVAΔC are capable of recognizing PA and PI3P, but to different extents. Dotted lipids are as follows: lysophosphatidic acid (LPA), lysophosphatidylcholine (LPC), phosphatidylinositol (PI), phosphatidylinositol (3)-phosphate (PI3P), phosphatidylinositol (4)-phosphate (PI4P), phosphatidylinositol (5)-phosphate (PI5P), phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine 1-phosphate (S1P), phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2), phosphatidylinositol (3,5)-bisphosphate (PI(3,5)P2), phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2), phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3), phosphatidic acid (PA), and phosphatidylserine (PS). (d) Glide induced fit docking results of the head groups of PA (left) and PI3P (right) against the PH domain of MINERVA. Sidechains of residues for lipid binding are indicated in stick and surface representations. Binding free energy for PA and PI3P headgroups calculated by the molecular mechanics–generalized Born surface area (MMGBSA) method is shown, respectively.