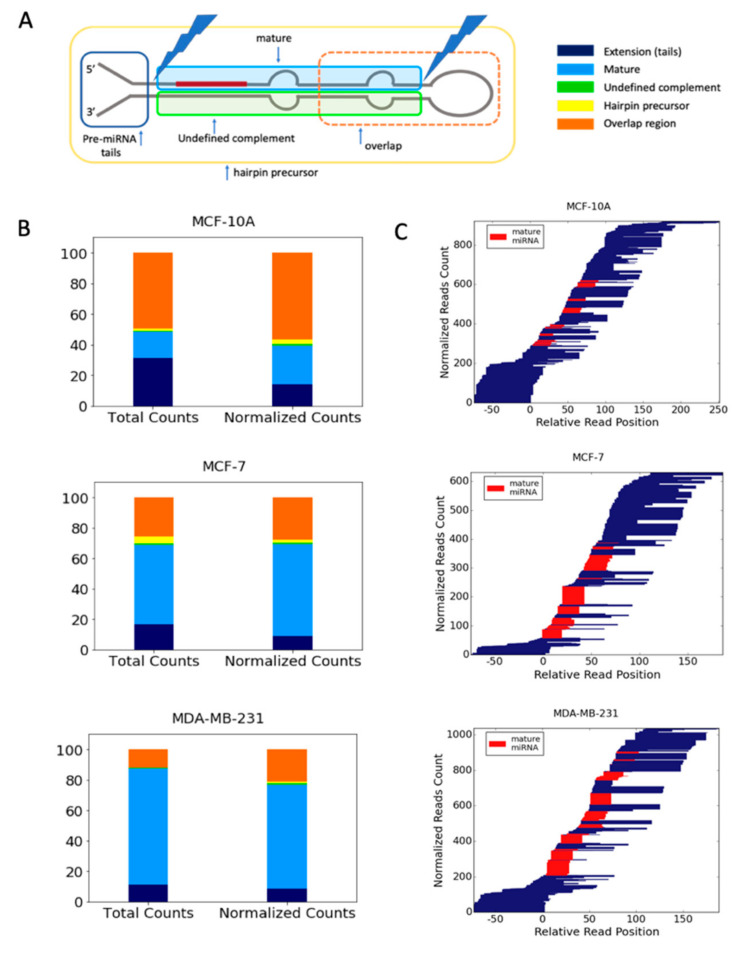
Figure 4.
Partition of the SF-miRNAs to segmental pre-miRNA regions. (A) A schematic view of an miRNA prototype, where the different regions of the HP-miRNA are listed according to their positions with respect to the pre-miRNA major processing sites. An example for an overlap region, defined as reads that cross known segmental boarders, is indicated. (B) The relative counts of reads mapped to each of the predefined regions for each of the three cell lines. The relative counts of reads to each of the pre-miRNA regions are color-coded as in A. Total counts of reads (left) and normalized counts (right, the relative abundance of reads aligned to a specific miRNA sums to 1) by each miRNA are shown for MCF-10A, MCF-7, and MDA-MB-231. (C) Blocks of reads from the same starting points reflect non-randomized cleavage sites. The 5p and 3p mature miRNAs are colored red, and the width of the bars is proportional to the number of reads. The position and alignment of the mature miRNAs is according to the miRbase annotations. All miRNAs that are analyzed have an average minimal number of reads per cell-line (≥3), and a minimal read length of ≥17. Data source is in Supplemental Table S4.