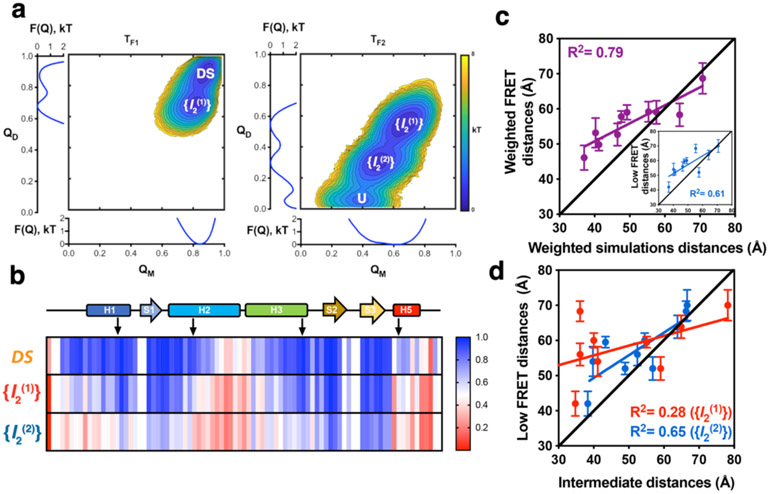
Figure 4. Structure-based MD simulations of FoxP1 dimer.
a) Folding free energy landscape of FoxP1 dimer, calculated from 1.6E8 timestep simulations in different temperature conditions, where TF1 and TF2 are the two peaks of the changes in specific heat as a function of temperature observed for FoxP1. Two clearly defined basins, DS and the intermediate state ({I2(1)}) can be seen at T = TF1. In contrast, three states were observed at T = TF2, corresponding to {I2(1)}, a second intermediate ensemble state ({I2(2)}), and the unfolded state (U). b) Changes in the per-residue probabilities of intramolecular native contact formation (from 0 to 1) of FoxP1 in the DS conformation and in the described dimeric intermediate ensembles {I2(1)} and {I2(2)}. c) Correlation between the weighted donor-acceptor distances determined via PDA and the weighted theoretical FRET distances estimated based on MD simulations. Inset: correlation between the low FRET distances determined by PDA and the weighted theoretical FRET distances estimated based on the MD simulations. d) Correlation between low FRET distances derived from PDA and the representative structure of {I2(1)} and {I2(2)}.