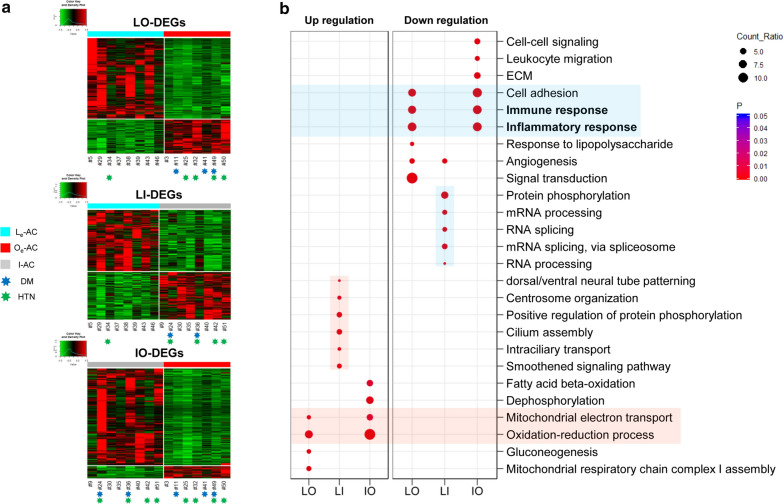
Fig. 3.
Three classes of DEGs using the three redefined groups of samples. a Construction of heatmap coupled with unsupervised hierarchical clustering for three different classes of DEGs. A total of 2,657 ‘LO-DEGs’ (Q < 0.01), 1,474 ‘LI-DEGs’ (P < 0.01), and 1,324 ‘IO-DEGs’ (Q < 0.05) are used to construct a heatmap coupled with unsupervised clustering (refer to Additional file 4: Table S3). The same notations used for Fig. 2a are also used in this heatmap (refer to Fig. 2a legend). b Analysis of GO functional terms for each of the three classes of DEGs. ‘LO’, ‘LI’, and ‘IO’ represent ‘LO-DEGs’, ‘LI-DEGs’ and ‘IO-DEGs’, respectively. For each class of DEGs, genes are divided into upregulated genes (i.e., genes with higher levels in obese ACs than in lean ACs) and downregulated genes (i.e., genes with lower levels in obese ACs than in lean ACs). Wide red and blue boxes within the plot indicate upregulated and downregulated genes, respectively, for both ‘LO-DEGs’ and ‘IO-DEGs’. Narrow red and blue boxes within the plot indicate upregulated and downregulated genes, respectively, for the ‘LI-DEGs’