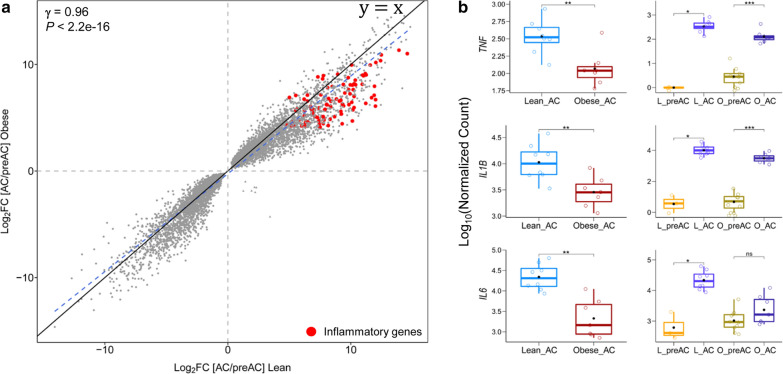
Fig. 6.
Comparison of inflammatory gene expression in lean and obese adipogenesis. a A scatter plot was constructed with a total of 7,118 genes commonly altered in lean and obese adipogenesis (i.e., common DEGs between ‘Lean_Ag-DEGs’ and ‘Obese_Ag-DEGs’). Log2FC values of gene expression in ‘Lean_Ag-DEGs’ were plotted against log2FC values of gene expression in ‘Obese_Ag-DEGs’. The gray and red dots represent the total 7118 common DEGs and the inflammatory genes, respectively. The blue dashed diagonal line indicates a regression line for the values, while the black diagonal line indicates the line of equality (y = x). The correlation coefficient (γ) was calculated by Pearson correlation test. b Boxplots of gene expression values for three selected genes, i.e., IL-6, TNF-α, and IL-1β were constructed; the box plots in the left panel provide a comparison of the expression of these three genes in lean and obese AC samples, while the box plots in the right panel provide a comparison between ‘[AC/preAC]_lean’ (i.e., expression levels of these three genes in AC and preAC samples derived from the lean adipogenesis condition) and ‘[AC/preAC]_obese’ (i.e., expression levels of these three genes in AC and preAC samples derived from the obese adipogenesis condition). The statistical test was performed by the ‘Wilcoxon rank-sum test’. ‘*’, ‘**’, and ‘***’ indicate P < 0.05, P < 0.01, and P < 0.001, respectively, while ‘ns’ indicates P > 0.05