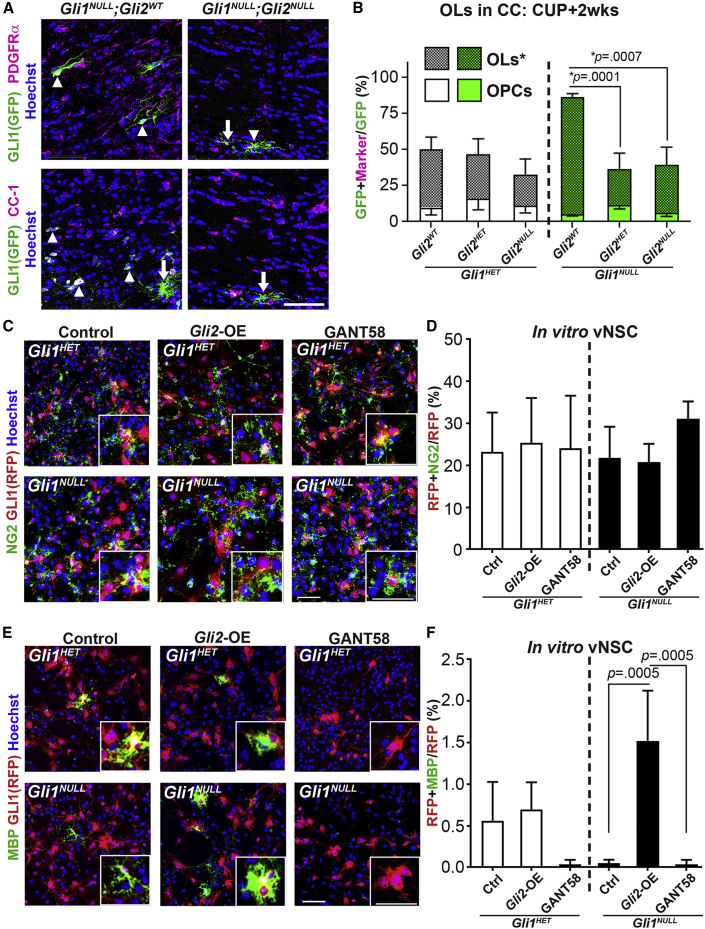
Figure 4.
Gli2 Promotes Oligodendrocyte Differentiation from vNSCs
(A) Immunofluorescence for fate-mapped Gli1NULL vNSCs (green) co-expressing PDGFRα (OPC) or CC1 (OL) (magenta). n = 5 mice/genotype, GFP + cell (arrows) and marker + GFP co-expressing cells (arrowheads).
(B) Quantification of the proportion of GFP + fate-mapped Gli1HET (white) and Gli1NULL (green) vNSCs that co-express markers of OPC (PDGFRα) and OL (CC1) in (B).
(C) Immunofluorescence for OPCs (NG2, green) and RFP + Gli1 fate-mapped vNSCs (red) following overexpression (Gli2-OE) or inhibition (GANT58 treatment) of Gli2 in vitro. n = 3 replicates.
(D) Quantification of Gli2 overexpression or inhibition in (C).
(E) Immunofluorescence for MBP + mature OLs (green) and RFP + fate-mapped Gli1 vNSCs (red) following overexpression (Gli2-OE) or inhibition (GANT58 treatment) of Gli2 in vitro. n = 3 replicates.
(F) Quantification of Gli2 overexpression (Gli2-OE) or inhibition (GANT58 treatment) in (E).
One-way ANOVA with Tukey's post-hoc t tests within Gli1 genotypes; all data presented as mean ± SEM. Scale bars, 50 μm. CC, corpus callosum; OPC, oligodendrocyte progenitor cells; OL, oligodendrocyte; CUP, cuprizone diet; Gli2-OE, Gli2-overexpression.