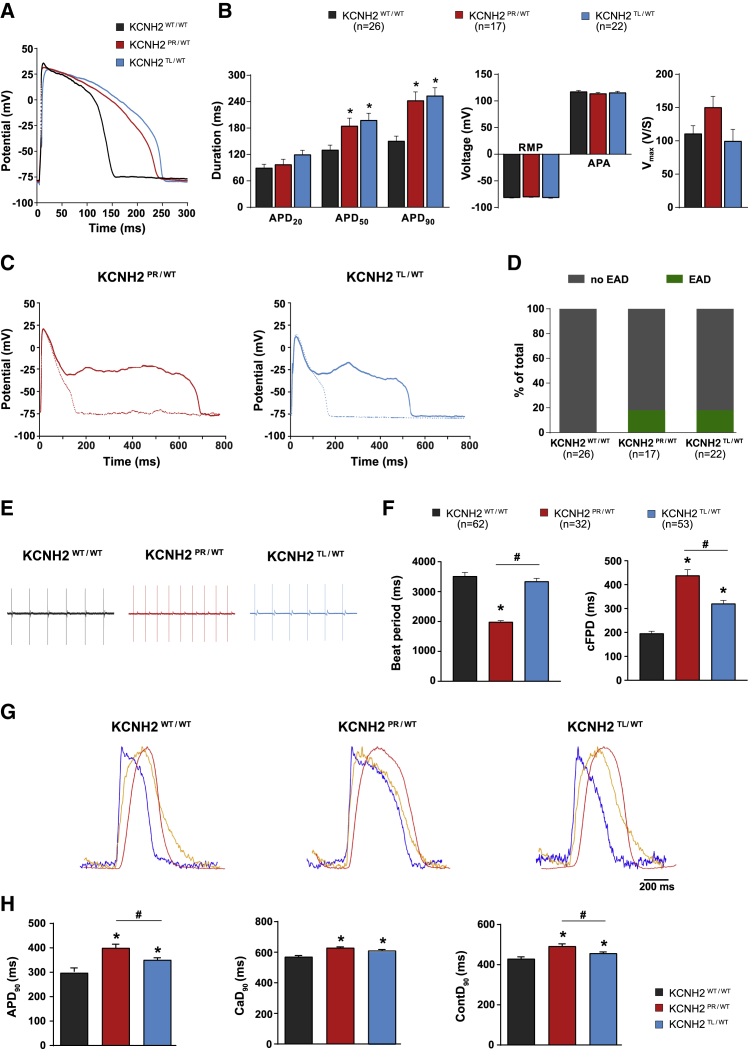
Figure 3.
Electrophysiological Characterization of the KCNH2WT/WT, KCNH2PR/WT, and KCNH2TL/WT hiPSC-CMs
(A and B) Representative AP traces (A), and average APD20, APD50, APD90, RMP, AP amplitude (APA), and Vmax values (B) for the indicated lines paced at 1 Hz. ∗Statistical significance to KCNH2WT/WT (KCNH2PR/WT: APD50, p < 0.05; APD90, p < 0.001; KCNH2TL/WT: APD50, p < 0.01; APD90, p = 0.0001; one-way ANOVA with Tukey's multiple comparisons test). Values (n) refer to the number of individual hiPSC-CMs analyzed.
(C) Consecutive AP traces from a KCNH2PR/WT (left) and KCNH2TL/WT (right) hiPSC-CM with and without (solid and dotted lines, respectively) oscillations in membrane potential interrupting the repolarization and indicating the occurrence of EADs.
(D) Percentage of the indicated lines displaying EADs when paced at 0.2 Hz. Values (n) refer to the number of individual hiPSC-CMs analyzed.
(E and F) Representative MEA traces (E) and average values for beat period and cFPD (F) for the indicated lines. ∗Statistical significance to KCNH2WT/WT (KCNH2PR/WT: beat period, cFPD, p < 0.0001; KCNH2TL/WT: cFPD, p < 0.0001; one-way ANOVA with Tukey's multiple comparisons test. #Significance between KCNH2PR/WT and KCNH2TL/WT (beat period, cFPD, p < 0.0001). Values (n) refer to the number of independent wells analyzed from at least four differentiations for each cell line.
G) Representative averaged time plots of baseline-normalized fluorescence signals for the indicated lines stimulated at 1.2 Hz. AP traces are shown in blue, cytosolic Ca2+ flux in orange, and contraction-relaxation in red.
(H) Average APD90, CaD90, and ContD90 values for the indicated lines as determined by changes in fluorescent signal. ∗Statistical significance to KCNH2WT/WT (KCNH2PR/WT: APD90, CaD90, ContD90, p ≤ 0.0001; KCNH2TL/WT: APD90, ContD90, p < 0.05; CaD90, p < 0.001); #statistical significance between KCNH2PR/WT and KCNH2TL/WT (APD90, ContD90, p < 0.05); n = 35–39 recordings from three differentiations for each cell line (Student's t test following one-way ANOVA with Tukey's multiple comparisons test).
Error bars represent SEM.
See also Figure S3.