Summary
The role of leptin receptor (OB-R) signaling in linking pluripotency with growth and development and the consequences of dysfunctional leptin signaling on progression of metabolic disease is poorly understood. Using a global unbiased proteomics approach we report that embryonic fibroblasts (MEFs) carrying the db/db mutation exhibit metabolic abnormalities, while their reprogrammed induced pluripotent stem cells (iPSCs) show altered expression of proteins involved in embryonic development. An upregulation in expression of eukaryotic translation initiation factor 4e (Eif4e) and Stat3 binding to the Eif4e promoter was supported by enhanced protein synthesis in mutant iPSCs. Directed differentiation of db/db iPSCs toward the neuronal lineage showed defects. Gene editing to correct the point mutation in db/db iPSCs using CRISPR-Cas9, restored expression of neuronal markers and protein synthesis while reversing the metabolic defects. These data imply a direct role for OB-R in regulating metabolism in embryonic fibroblasts and key developmental pathways in iPSCs.
Keywords: leptin receptor, pluripotency, embryonic development, diabetes, cancer, neuronal lineage, STAT3, EIF4E
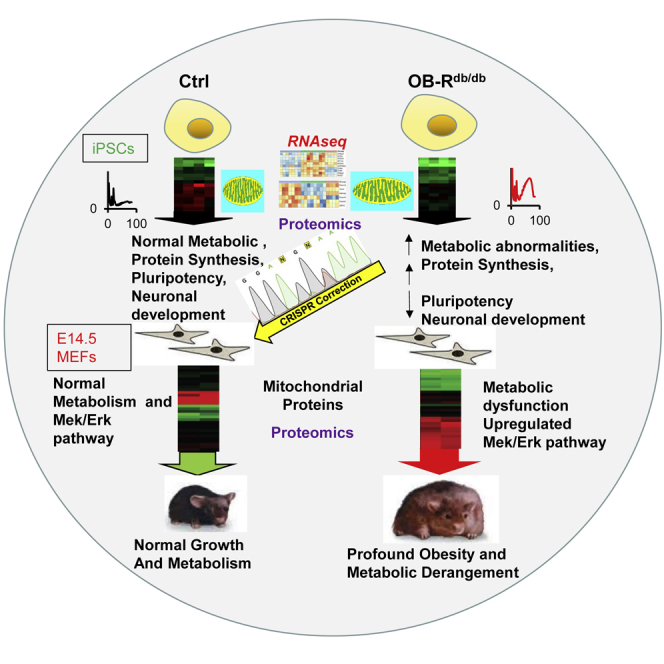
Graphical Abstract
Highlights
-
•
Pluripotency markers are decreased in db/db iPSCs (lacking functional OB-R)
-
•
Mouse db/db iPSCs exhibit higher protein synthesis mediated by the Stat3/Eif4e axis
-
•
OB-R signaling regulates neuronal development markers—NOGGIN, NESTIN, GFAP
-
•
CRISPR correction reverses defects in db/db iPSCs
In this article Kulkarni and colleagues use CRISPR-Cas9 gene editing and pluripotent stem cells as a model system to report that leptin receptor regulates protein synthesis pathways, pluripotency markers, and early neural development in early stages of life.
Introduction
Mutations in different exons in the leptin receptor (OB-R) gene have been reported to cause obesity, pituitary dysfunction, and metabolic abnormalities in humans (Clement et al., 1998; Saeed et al., 2014). The db/db mouse, which harbors a mutation in the OB-R gene (G to T transition in the intron region between exon 18 and exon 19), manifests features that are strikingly similar to those seen in humans carrying the mutant receptor, such as obesity, type 2 diabetes, and tumorigenesis (Chen et al., 1996; Kobayashi et al., 2000; Wang et al., 2014). Thus, the db/db mouse continues to be widely used as a suitable model to explore the role of OB-R in the development and progression of obesity, diabetes, and its related complications.
While a majority of circulating leptin originates from adipose tissue, other tissues also contribute, such as the placenta, ovaries, testes, skeletal muscle, cartilage, bone cells, and stomach (Margetic et al., 2002). Following secretion leptin acts mainly via the OB-R, of which several isoforms exist, differing mainly in the length of the cytoplasmic domain (Chen et al., 1996). In addition to expression in adult tissues, such as the heart, pancreatic beta-cells, and immune cells the OB-R has been detected in fetal and developmental tissues, including placenta, endometrium, and pluripotent stem cells (El-Hefnawy et al., 2000; Hoggard et al., 2001). Furthermore, leptin, secreted by fibroblasts, has been reported to support proliferation of human pluripotent stem cells (Anisimov et al., 2011) and a previous study demonstrated binding of pluripotency factors, Oct4 and Sox2, on the promoter region of OB-R (Feldman et al., 2012). Recently, leptin signaling has been suggested to regulate the parasympathetic nervous system during early embryonic development (Croizier et al., 2016). Thus, while studies point to a role for leptin receptors during early development the direct functional role of OB-R in modulating pluripotency and its consequences on development is unknown.
In this study, we used mouse embryonic fibroblasts (MEFs) and iPSCs derived from db/db mice as complementary model systems to explore OB-R signaling in the regulation of pluripotency and metabolic/mitochondrial function during embryonic development. Strikingly, db/db MEFs exhibited an upregulated ERK pathway and metabolic dysfunction, while db/db iPSCs showed reduced pluripotency and differential regulation of protein synthesis pathways. The latter defects in iPSCs were reversed upon correction of the db/db point mutation by genome editing using the clustered regularly interspaced short palindromic repeats (CRISPR-Cas9) approach. These findings were supported by RNA sequencing (RNA-seq) analyses on iPSCs. Furthermore, we report a role for OB-R signaling during neuronal development. Together these data indicate the significance of the OB-R in regulating genes/proteins relevant for pluripotency, protein synthesis pathways, and neuronal development with potential implications for cancer and metabolic diseases.
Results
Functional Characterization and Global Proteomics Reveal Metabolic Abnormalities in Embryonic db/db MEFs
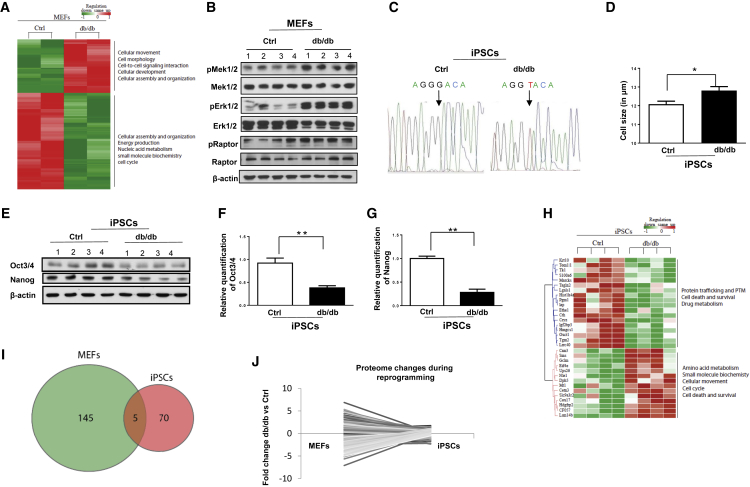
Global differential protein expression between Ctrl and db/db MEFs, investigated by mass spectrometry-based proteomics, revealed 38 strongly and 92 moderately regulated proteins that were significantly different between groups (detailed in the Experimental Procedures). The most differentially regulated proteins between Ctrl and db/db MEFs were those related to mitochondrial function and oxidative phosphorylation. The differentially regulated proteins and pathways are listed in Table S1 (Figures 1A, S1A, and S1B).
Figure 1.
Altered Metabolic Regulation in db/db MEFs and Reduced Pluripotency and Upregulated Protein Synthesis Pathway in db/db iPSCs
(A–G) (A) Heatmap representation of differentially expressed proteins between Ctrl and db/db MEFs. A selection of gene ontology terms is given for each protein cluster. Red indicates upregulated while green indicates downregulated proteins in db/db MEFs. (B) Western blots analysis showing upregulated MEK1/2, ERK1/2, and RAPTOR pathways in db/db MEFs. β-Actin was used as a loading control. (C) Genomic DNA sequencing showing G to T transition of base in db/db iPSCs. (D) Cell diameter of Ctrl and db/db iPSCs using Cello meter. (E) Western blot analysis of OCT4 and NANOG in four different clones from Ctrl and db/db iPSCs; quantification of changes in OCT4 (F) and NANOG (G) using ImageJ software. β-Actin was used as a loading control.
(H–J) (H) Heatmap representation of pathway analysis of differentially expressed proteins between Ctrl and db/db iPSCs. A selection of GO terms is given for each protein cluster. Red indicates upregulated while green indicates downregulated proteins in db/db iPSCs. (I) Venn diagram showing the numbers of proteins that were significantly differently expressed (p < 0.05) in db/db MEF-specific proteome and the db/db iPSC-specific proteome. Five proteins were significantly changed between db/db and Ctrl, both before and after reprogramming. (J) Pairing of protein expression values from each of the significant proteins in the db/db MEF-specific proteome with the db/db iPSC-specific proteome.
The most differentially regulated pathways included mitochondrial dysfunction, oxidative phosphorylation, and integrin signaling, while the moderately regulated pathways were insulin growth factor 1 signaling, cell cycle, BMP signaling, DNA methylation, molecular mechanisms of cancer, adipogenesis, leptin signaling in obesity, and protein ubiquitination. The complete list of regulated pathways is presented in Table S2.
To validate the key pathways we began with morphological characterization and observed larger MEFs in the db/db group (Figure S1C). We observed an upregulation of phosphorylation of MEK1/2, ERK1/2, and RAPTOR in db/db MEFs (Figure 1B), and the abundance of CDKN2a was upregulated in these MEFs (Figure S1D). Together, these data point to differential regulation of the MEK/ERK/RAPTOR pathway and mitochondrial dysfunction in db/db MEFs during embryonic development and suggest a link between OB-R and obesity and oncogenesis.
db/db iPSCs Show Altered Morphology and Pluripotency Characteristics
We confirmed the persistence of the leptin receptor mutation (G to T transition) in the db/db group by sequencing genomic DNA (Figures 1C and S2A). While no difference in colony morphology was evident between the two groups, a significantly larger cell size was detected in db/db iPSCs (Figures 1D and S2B). Interestingly, western blot analysis showed significant downregulation of Oct4 and Nanog protein levels in db/db iPSCs (Figures 1E–1G).
To explore their ability to spontaneously differentiate in vivo into the three germ layers we injected Ctrl or db/db iPSCs (1 × 106 cells/mouse) either subcutaneously or intramuscularly into severe combined immunodeficient (immunodeficient) and B6 (syngeneic) mice. As expected, we observed teratomas in both groups and the presence of β-III-tubulin (ectoderm in green), α-actinin (mesoderm in green), and HNF-3-β (endoderm in red) (Figures S2C and S2D) confirmed components of the three germ layers.
Global Proteomics Analyses of Ctrl and db/db iPSCs Reveals Differentially Regulated Pathways Associated with Pluripotency and Protein Synthesis
To interrogate changes in the global proteome we undertook systematic comparisons between MEFs and iPSCs separately in Ctrl and db/db groups as well as Ctrl iPSCs versus db/db iPSCs. We identified 665 strongly regulated proteins and 278 moderately regulated proteins that were significantly different between the two cell types (Ctrl MEFs versus Ctrl iPSCs) (Table S3). A comparison between db/db MEFs and db/db iPSCs, revealed 789 strongly regulated proteins and 271 moderately regulated proteins that reached statistical significance (Table S4). Comparing the proteome between Ctrl and db/db iPSCs, we identified 32 significantly and moderately regulated proteins (Table S5).
Next, to assess the specific effects of leptin signaling at the proteome level in pluripotent stem cells, we aimed to identify a protein signature consequent to deregulated OB-R signaling in induced pluripotent stem cells derived from db/db mice. The heatmap analyses revealed that mitochondrial proteins, protein synthesis, and pluripotent pathways were differentially regulated between Ctrl and db/db iPSCs (Figures 1H and S3A). As expected, iPSCs from Ctrl and db/db showed fewer differences in proteomic changes than the differentiated MEFs from the two respective groups. Specifically, up to 150 proteins were significantly differently expressed in the db/db MEF-specific proteome compared with 75 in the db/db iPSC-specific proteome, with five proteins being significantly differently expressed in both proteomes (Figure 1I).
Notably, many of the highly regulated proteins in the db/db MEF-specific proteome did not persist in the iPSC-specific proteome, suggesting a shutdown or suppression of db/db-specific proteins and pathways after the MEF cells were reprogrammed into pluripotent cells (Figures 1I and 1J).
Upregulated Eukaryotic Translation Initiation Factor 4E Protein Synthesis Pathway and Higher Metabolic Rates in Db/db iPSCs
Proteomic analyses revealed an upregulation of the eukaryotic translation initiation factor 4e (EIF4E) protein synthesis pathway among the canonical networks in the iPSCs of db/db mice. The complete list of differentially regulated pathways is available in Table S2 (Figures S3A and S3B; Tables S2 and S5).
We next considered previous reports that tissues manifesting the db/db mutation exhibit altered signal transducer and activator of transcription 3 (STAT3) signaling. Interestingly, while the effects of the db/db mutation have been observed in the hypothalamus (Ghilardi et al., 1996) leptin has been reported not to activate the Stat pathway in other adult tissues (Vaisse et al., 1996). The latter prompted us, for the first time, to examine STAT3 activation in iPSCs. In the unstimulated state, Ctrl iPSCs showed higher phosphorylation of STAT3, which robustly increased after stimulation with leukemia inhibitory factor (LIF) or leptin. The mutant db/db iPSCs showed a similar level of phosphorylation of STAT3 as compared with Ctrl iPSCs in the unstimulated state. Although db/db iPSCs also demonstrated phosphorylation of STAT3 after stimulation with LIF, the effect was significantly blunted after leptin stimulation indicating defective signaling (Figure S3C). Furthermore, OCT4 protein abundance was decreased in db/db iPSCs in the starved condition similar to that in the basal state.
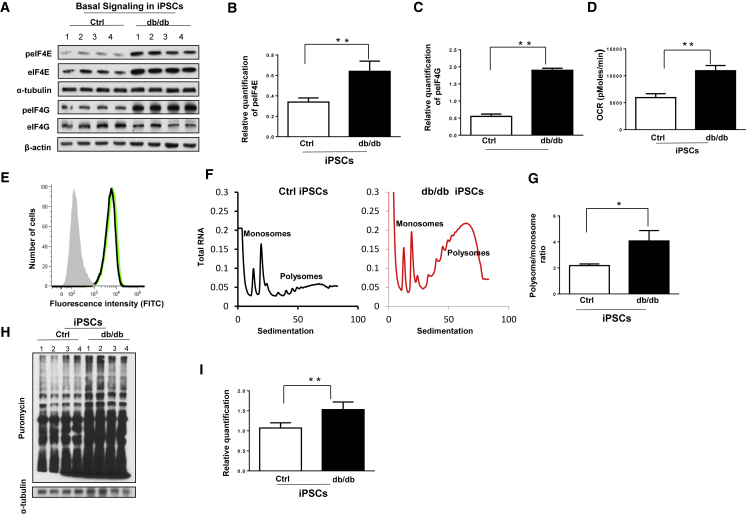
EIF4E, as well as phosphorylated EIF4E, were significantly upregulated in the basal state, in agreement with mass spectrometry data on db/db iPSCs (Figures S3A and S3B). Phosphorylated EIF4G was also significantly upregulated in db/db iPSCs in the basal state as compared with Ctrl iPSCs (Figures 2A–2C). These results indicate that disrupted leptin receptor signaling leads to higher abundance of proteins that regulate protein synthesis in the pluripotent stage.
Figure 2.
Upregulated Protein Synthesis in db/db iPSCs
(A–E) (A) Western blot analysis showing increase in phospho-EIF4E, phopho-EIF4G, and total EIF4E in db/db iPSCs. (B and C) quantification of the data in (A) phospho-EIF4E (B) and phospho-EIF4G (C). (D) metabolic profiling by Seahorse showing higher oxygen consumption rate (OCR) in db/db iPSCs as compared with Ctrl iPSCs. (E) Flow cytometry analysis showing relative higher mitochondrial mass (Mito-GFP) in db/db iPSCs.
(F–I) (F) Representative polysome profiling graphs showing polysome and monosome peaks in Ctrl (left panel, in black) and db/db iPSCs (right panel, in red). (G) Quantification of polysome and monosome ratios in both groups. (H) Anti-puromycin western blot analysis demonstrated higher protein synthesis in db/db iPSCs as compared with Ctrl iPSCs. α-Tubulin was used as a loading control. (I) Quantification of newly synthesized proteins in Ctrl and db/db iPSCs using α-tubulin as a loading control. n = 3 independent experiments; data are shown as mean ± SD by Student's t test (∗p < 0.05, ∗∗p < 0.01).
Next, metabolic profiling revealed higher basal oxygen consumption rate (OCR) in db/db iPSCs, indicating an increased metabolic rate/oxidative phosphorylation (OXPHOS) compared with Ctrl iPSCs (Figure 2D). Electron microscopy and flow cytometry analysis revealed relatively higher mitochondrial numbers in db/db iPSCs (Figures 2E and S4A). There was also a significant upregulation in the expression of Pgc-beta and G6pd genes in db/db iPSCs as compared with Ctrl iPSCs (Figures S4B and S4C). Together, these data suggest that the presence of the db/db mutation is associated with higher metabolic activity in the iPSCs.
Polysome Profiling Reveals Higher Protein Synthesis in db/db iPSCs: Role of Stat3 Binding to eIF4E Promoter
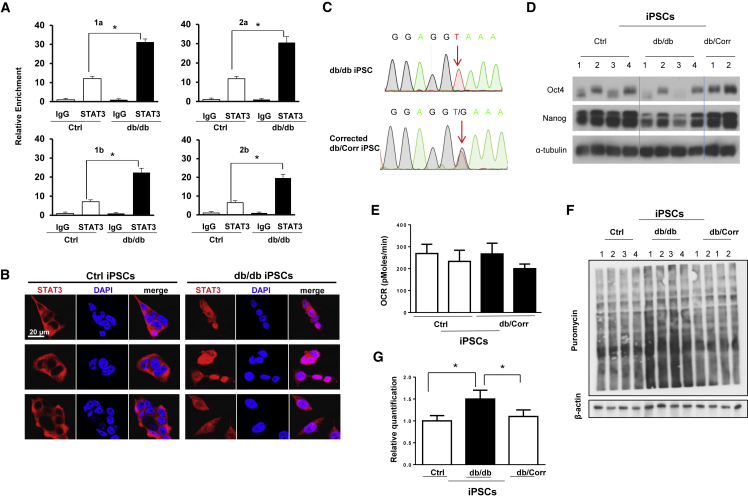
Since proteins related to protein synthesis were upregulated in db/db iPSCs, we undertook polysome profiling and observed a higher polysome/monosome ratio in db/db iPSCs suggesting enhanced protein synthesis (Figures 2F and 2G). These data were confirmed by an alternative non-radioactive approach (Schmidt et al., 2009) wherein addition of puromycin to a growing culture of Ctrl or db/db iPSCs led to increased protein synthesis in the latter at the end of 48 h (Figures 2H and 2I). In silico analysis revealed two Stat3 binding regions on the promoter of the Eif4e gene, which might be involved in cellular protein synthesis (Figure S5A). Indeed, chromatin immunoprecipitation (ChIP) analysis by real-time PCR, to validate binding of Stat3 to the promoter of Eif4e, showed significantly greater enrichment of Stat3 binding on the Eif4e promoter region in db/db iPSCs as compared with Ctrl iPSCs. The relative enrichment of two Stat3 binding regions (region 1: from −841 to −834; region 2: from −168 to −160) on the promoter of Eif4e were achieved by using 2 pairs of primers to examine each region (1a, 1b for “GGCTTCCC” and 2a, 2b for “TTCCCAGAA”) in real-time PCR analysis (Figures 3A and S5B). Moreover, live immunostaining showed predominantly nuclear localization of STAT3 in db/db iPSCs even in the basal state (Figure 3B). Together, these results implicate a role for Stat3 in the regulation of protein synthesis via Eif4e in db/db iPSCs.
Figure 3.
Chromatin Immunoprecipitation Analysis for Stat3-Eif4e Binding and CRISPR Correction of the db/db Mutation in iPSCs
(A–E) (A) Chromatin immunoprecipitation analysis showing higher Stat3 binding (relative enrichment) on the two promoter regions “a” and “b” of Eif4e in db/db iPSCs using four different sets of primers 1a, 1b and 2a, 2b. (B) Live-confocal microscopy reveals the predominant nuclear localization of STAT3 in db/db iPSCs compared with cytosolic localization of STAT3 in Ctrl iPSCs. Scale bar, 20 μm. Each row indicates a single clone from Ctrl and db/db iPSCs. (C) db/db mutation (G to T) in the intronic region between exon 18 and exon 19, and DNA sequencing reveals the reversal of one copy to wild type in db/db iPSCs. Similar results were obtained in two additional clones. (D) Western blot analysis showing restoration of protein abundance of OCT4 and NANOG in CRISPR-corrected (db/Corr) iPSCs. α-Tubulin was used as a loading control. (E) Metabolic profiling showed similar basal respiration in db/db-corrected iPSCs compared with Ctrl iPSCs.
(F and G) (F) Anti-puromycin western blot analysis showing normalization of protein synthesis in CRISPR-corrected db/Corr iPSCs. β-Actin was used as a loading control. (G) Quantification of data from experiment in (F), n = 3 independent experiments; data are shown as mean ± SD and analyzed by Student's t test (∗p < 0.05, ∗∗p < 0.01).
CRISPR-Cas9 Correction of db/db iPSCs Normalizes Altered Protein Synthesis and Pluripotency Pathways
To determine whether the mutation in the OB-R is directly relevant for the phenotypes observed in the iPSCs, we designed guide RNAs to target and correct the G to T point mutation using CRISPR-Cas9 genome editing. A control template with the wild-type sequence was used to replace the mutation (Figure S6A). Among the 192 clones selected for sequencing we identified two clones with one copy reverted to the wild-type sequence (db/+), hereafter termed db/Corr iPSCs (Figures 3C and S6B). To examine the effect of the db/db mutation, we performed molecular and functional analysis of Ctrl and CRISPR-corrected (db/Corr) iPSCs. Western blot analysis showed that db/Corr iPSCs exhibited EIF4E and OCT4 protein expression that was similar to Ctrl iPSCs in the basal state (Figures 3D and S6C). Furthermore, metabolic profiling demonstrated similar levels of OCR in the mutation db/Corr iPSCs and Ctrl iPSCs (Figure 3E). Anti-puromycin antibody-based Western blot analyses also revealed that protein synthesis in CRISPR-corrected db/Corr iPSCs is not significantly different from Ctrl iPSCs (Figures 3F and 3G).
RNA-Seq Analyses of iPSCs Authenticate the Role of OB-R in Protein Synthesis and Pluripotency Pathways
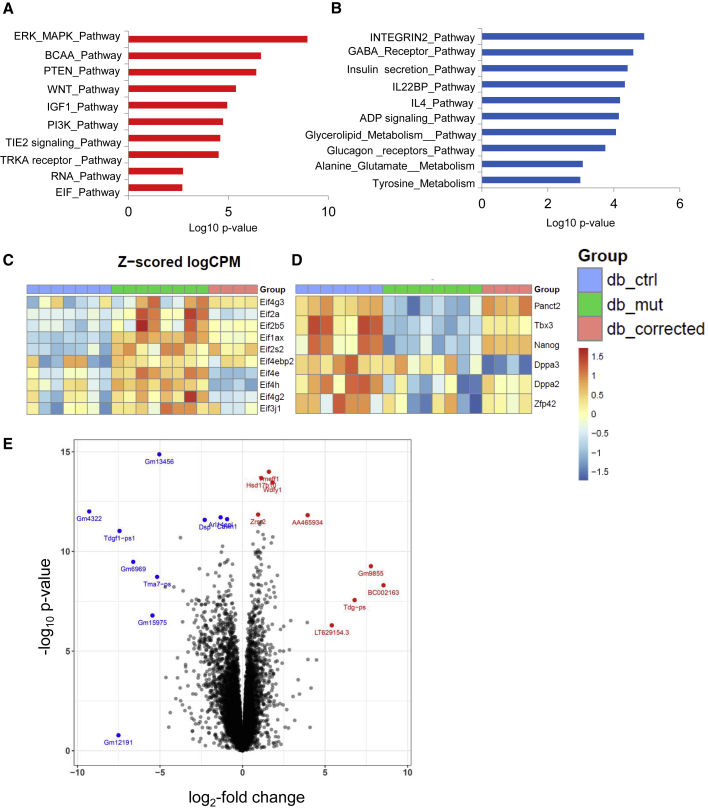
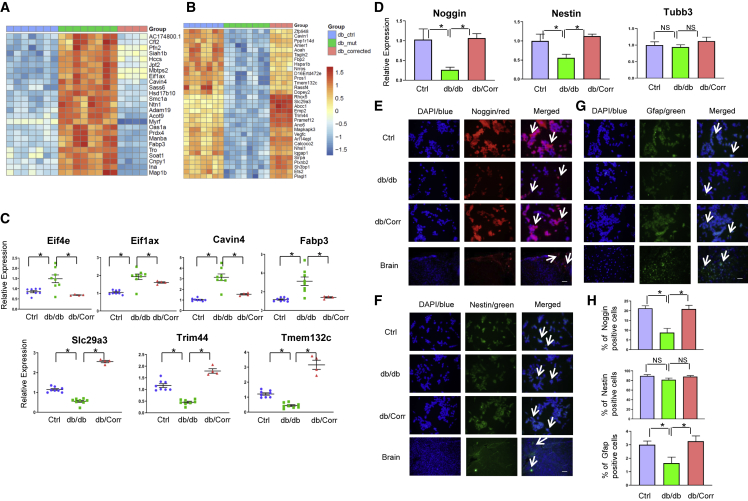
To confirm our previous findings, we conducted RNA-seq analyses on Ctrl, db/db, and db/Corr iPSCs. A total of 1,662 upregulated and 1,887 downregulated genes were determined between Ctrl and db/db iPSCs with false discovery rate < 0.25 and adjusted p value < 0.05. This included 651 upregulated genes and 630 downregulated genes between db/db versus Ctrl and db/Corr versus Ctrl iPSCs. Gene ontology and pathway analyses between Ctrl and db/db iPSCs showed an upregulation in the ERK and EIF pathways and downregulation in insulin secretion, alanine-glutamate metabolism, and tyrosine metabolic pathways (Figures 4A and 4B). Heatmaps confirmed the proteomics data with an upregulation of genes related to the protein synthesis machinery (Figures 4C and 5A), while pluripotency-associated genes were downregulated (Figures 4D and 5B) in db/db iPSCs. These alterations were reversed in db/Corr iPSCs.
Figure 4.
RNA-Seq Analyses Confirms Upregulation of Protein Synthesis Pathways and Downregulation of Pluripotency-Associated Genes
(A–C) Red color indicates upregulated pathways (A) and blue color indicates downregulated pathways (B) in db/db iPSCs as compared with Ctrl iPSCs. (C) Several genes related to protein synthesis pathways were upregulated in db/db iPSCs and were normalized after CRISPR-Cas9 correction of iPSCs.
(D) Pluripotency genes were downregulated in db/db iPSCs.
(E) Volcano plot showing differentially regulated genes between Ctrl and db/db iPSCs. Red color indicates genes that are upregulated and blue color indicates downregulated genes in db/db iPSCs as compared with Ctrl iPSCs. n = 3 independent experiments; data are shown as mean ± SD and analyzed by Student's t test (∗p < 0.05, ∗∗p < 0.01).
Figure 5.
Neuronal Differentiation of Ctrl, db/db, and db/Corr iPSCs Points to a Role for Leptin Receptors in the Regulation of Early Neuronal Markers
(A) Heatmap analyses showing upregulation of top differentially regulated genes in db/db iPSCs that were normalized in db/Corr iPSCs.
(B) Heatmap analyses showing downregulation of top differentially regulated genes in db/db iPSCs that were normalized in db/Corr iPSCs. Genes in red color denote upregulation while genes in blue color represent downregulation. Pathway analyses were conducted using Limma pathway software.
(C) Real-time PCR analyses of Eif4e, Eif1ax, Cavin4, Fabp3 (upregulated in db/db iPSCs), and Slc29a3, Trim44, Tmem132c (downregulated in db/db iPSCs) as a comparison with Ctrl iPSCs. Expression of these genes was normalized in corrected db/Corr iPSCs. β-Actin was used as a loading control. Gene expression in blue indicates Ctrl iPSCs, in green indicates db/db iPSCs, and in red indicates corrected db/Corr iPSCs.
(D) Real-time PCR analyses of early neuronal markers Noggin, Nestin, and Tubb3 in total RNA from day 15 neuronal cells.
(E–G) Representative images of Ctrl, db/db, and db/corr iPSC-derived neuronal cells (day 15) immunostained for markers of neuronal cells (E) NOGGIN (in red), (F) NESTIN (in green), and (G) GFAP (in green). All nuclei were stained in blue with DAPI. Mouse brain tissue was used as positive control. Scale bar, 100 μm.
(H) At least four independent images of each marker were quantified by manual counting. The percent of positive cells of each neuronal marker was determined by counting approximately 1,000 nuclei/image. n = 3 independent experiments; data are shown as mean ± SD and analyzed by Student's t test (∗p < 0.05, ∗∗p < 0.01).
Volcano plots between db/db versus Ctrl iPSCs revealed the top differentially regulated genes of OB-R in the pluripotent state (Figure 4E; Table S6). Validation studies using real-time PCR analyses confirmed the changes in several genes, including an upregulation of Eif4e, Eif1ax, caveolae-associated protein 4 (Cavin4), and fatty acid binding protein 3 (Fabp3), and downregulation of solute carrier family 29, member 3 (Slc29a3), tripartite motif-containing 44 (Trim44), and transmembrane protein 132c (Tmem132c) genes in db/db iPSCs. Again, the differential regulation was normalized in db/Corr iPSCs (Figure 5C).
Directed Differentiation of iPSCs Reveals a Role for OB-R Signaling in Development of the Neuronal Lineage
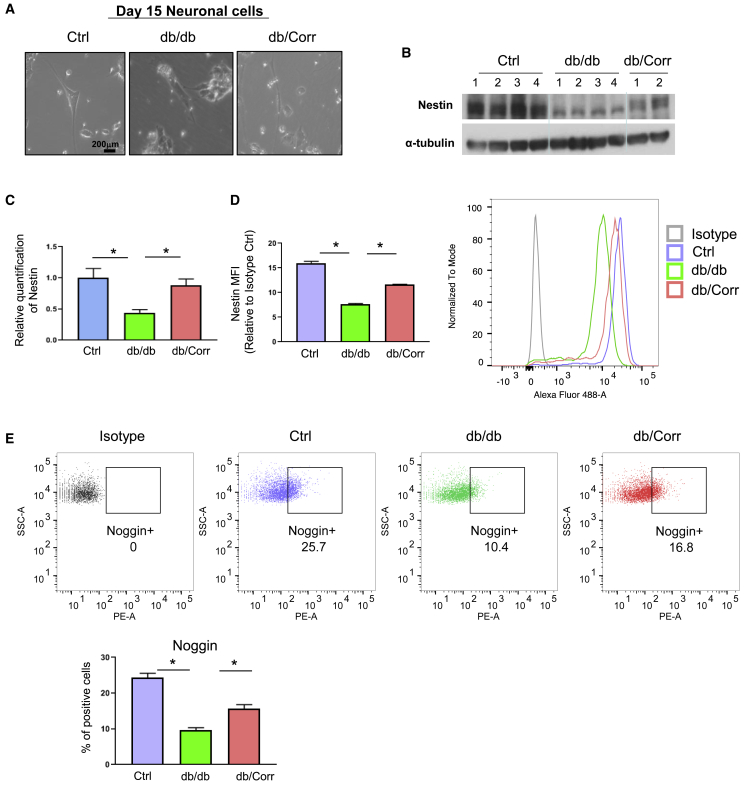
To investigate the role of leptin receptor signaling in tissues originating from the ectoderm we directed the differentiation toward the neuronal lineage (Gupta et al., 2018; Ying et al., 2003). After differentiation of the iPSCs, we observed downregulation of the neural progenitor markers Noggin and Nestin, while Tubb3+ did not show significant changes at the transcript level on day 15 in the db/db as compared with Ctrls (Figure 5D). Immunostaining analyses confirmed the decrease in NOGGIN+ cells (58.5%; p < 0.0001, n = 4) and glial fibrillary acidic protein (GFAP)+ cells (45.5%; p = 0.0019, n = 4) in db/db compared with Ctrl-derived neuronal cells. NESTIN+ cells were decreased in the db/db iPSC-derived neurons (5.4%, p = 0.07, n = 3) as compared with Ctrl neurons (Figures 5E–5H). Morphological images were taken at day 15 differentiated neuronal cells from Ctrl, db/db, and db/Corr iPSCs (Figure 6A). Despite a relatively small decrease in cell numbers we confirmed a decrease in NESTIN protein in db/db neuronal cells by western blot (∼45%; p < 0.05, n = 4) and flow cytometry analysis (52.1%; p < 0.0001, n = 3; decrease in mean fluorescent intensity) as compared with Ctrl neurons (Figures 6B–6D). The decrease in NOGGIN+ cells was also confirmed by flow cytometry analysis (60.3%; p < 0.0001, n = 3) in db/db neurons as compared with Ctrl neurons (Figure 6E). Importantly, the changes in expression of neuronal markers were normalized in db/Corr neurons that were differentiated after correction using CRISPR-Cas9. These results provide direct evidence that defects in leptin receptor signaling prompts mouse iPSCs to reduced/delayed differentiation toward the ectodermal lineage.
Figure 6.
Neuronal Differentiation of db/Corr iPSCs Shows the Ability of Gene Editing to Restore the Defects in Regulation of Early Neuronal Markers
(A) Morphological images of cells on day 15 of directed differentiation of Ctrl, db/db, or db/+ iPSCs. Scale bar, 200 μm.
(B) Total protein was isolated from day 15 differentiated neuronal cells. NESTIN antibody was used to detect its abundance in day 15 neuronal cells from Ctrl, db/db, or db/Corr groups. α-Tubulin was used as housekeeping control as shown in representative western blot.
(C and D) (C) Quantification of NESTIN western blot using ImageJ software. (D) NESTIN mean fluorescent intensity measurement of Ctrl, db/db, and db/Corr iPSC differentiated neuronal cells by flow cytometry.
(E) Flow cytometry analyses of NOGGIN+ cells from differentiated neuronal cells of Ctrl, db/db, and db/Corr iPSCs.
n = 3 independent experiments; data are shown as mean ± SD and analyzed by Student's t test (∗p < 0.05, ∗∗p < 0.01).
Discussion
db/db mice that manifest impaired leptin signaling are widely used as a disease model (Bates et al., 2005; Ernst et al., 2013). These homozygous mutant mice are born normally but develop diabetes and metabolic abnormalities at ∼6–8 weeks of age characterized by mitochondrial dysfunction (Holmstrom et al., 2012; Wang et al., 2014) while the heterozygous mice are relatively normal (Kobayashi et al., 2000). We report, for the first time, metabolic dysfunction and upregulated MEK/ERK pathways in the db/db embryonic fibroblasts that were morphologically larger and exhibited greater glycolytic and OX/PHOS capacities.
We used a proteomics approach and confirmed altered expression of OCT4 and EIF4E protein synthesis pathways in the db/db iPSCs. These observations have significant implications since OCT4 and NANOG are known to play key roles in maintaining pluripotency of iPSCs (Boiani et al., 2002; Chambers et al., 2003) and a quantitative change in the expression of Oct4 leads to differentiation, de-differentiation, or self-renewal of iPSCs (Niwa et al., 2000). Interestingly, RNA-seq analyses demonstrated that the expression of Nanog, Tbx3, Panct2, and Zfp42 were all decreased in db/db iPSCs, while the Oct4 transcript levels did not change. One interpretation of the latter observation is that OB-R is involved in post-transcriptional regulation of OCT4 at the pluripotent stage. Nevertheless, the reduced abundance of OCT4, NANOG, and other pluripotency markers in the db/db iPSCs suggests a potential impact on the differentiation process.
The translation initiation factor, eIF4e, has been reported to play an important role in protein synthesis and cell growth (Gingras et al., 1999; Sonenberg and Gingras, 1998). Our data indicate that disruption of OB-R leads to enhanced protein synthesis likely due persistent activation of Stat3 binding to the promoter of Eif4e in db/db iPSCs as shown by ChIP analysis and immuno-staining. While these observations may appear contradictory to previous reports of leptin stimulating protein synthesis in human trophoblast cells (Perez-Perez et al., 2009), and leptin acting via the OB-R to promote Stat3 phosphorylation and nuclear translocation in tumor initiating stem cells (Feldman et al., 2012), it is possible that leptin-activated Stat3 has context-dependent effects during the pluripotent stage. Furthermore, one cannot exclude the possibility of MEK/ERK pathways in activating EIF4E protein synthesis, similar to the effect of leptin on human trophoblast cell lines, in db/db iPSCs.
The successful restoration of metabolic function, pluripotency genes, and protein synthesis after correction of the db/db mutation (G to T) on one allele in db/Corr iPSCs using CRISPR-Cas 9 genome editing (Hsu et al., 2014) confirms a direct role for OB-R in maintenance of pluripotency and cell differentiation processes. The RNA-seq data on iPSCs confirms the observations after gene correction of OB-R with normalization of expression of several genes including Eif4e, Eif1ax to the level observed in Ctrl iPSCs.
Consistent with the report of leptin signaling being linked to neuronal embryonic development (Croizier et al., 2016), we observed a decrease in NOGGIN and GFAP+ cells and reduction in NESTIN protein, on day 15 of neuronal differentiation of db/db iPSCs. These changes were normalized in neuronal cells obtained from CRISPR-corrected db/Corr iPSCs. Mice lacking NOGGIN have been reported to develop defects in hypothalamic patterning, which is important for maintaining energy homeostasis during adulthood (Davis and Camper, 2007). Thus, in this scenario, and based on our findings, it is plausible that a decrease in NOGGIN+ cells in db/db mice during development leads to impaired development of POMC and AgRP neurons with consequences on development of obesity, diabetes, and metabolic syndrome as the mouse attains adulthood. Nestin has been reported to be important for self-renewal of neural progenitor cells and axonal growth of neurons (Bott et al., 2019) and its deficiency has been linked to impaired motor coordination (Mohseni et al., 2011). In our study, the reduction in Nestin protein could affect the neuronal projections/axonal growth that would in turn lead to impairment of normal neuronal development and coordination during adulthood.
Notably, previous studies by Ramos-Lobo et al. (2019) reported reduced Gfap expression in the hypothalamus of LepR null mice during early life with long-term consequences. Another study reported that modulation of Gfap-expressing glial cells in the hypothalamus is able to regulate feeding (Chen et al., 2016). In the context of these reports, the reduction in GFAP+ cells in db/db neuronal cells, as compared with Ctrl and db/Corr neurons, in our own study, argues for a potential reduction in GFAP+ glial cells in the hypothalamus with implications for feeding behavior. Our studies also gain significance since metabolic defects in the db/db mice, in contrast to the ob/ob model, cannot be fully restored by treatment with exogenous leptin (Pellymounter, 1995; Harris, 2001). Taken together, our findings indicate that normal leptin receptor expression and signaling regulates neural stem cell markers (e.g., NOGGIN and NESTIN) and GFAP protein during early developmental stages, which is necessary for maintenance of normal structural and/or functional properties of neurons populating the hypothalamus with implications for development of metabolic disorders in adulthood.
In summary, we report defects in MEFs and iPSCs derived from db/db mice with several implications. First, the regulation of Nanog, Tbx3, and Zfp42 at the transcript level and OCT4, and NANOG at the protein level in cells with the db/db mutation suggests a role for leptin signaling in regulation of “stemness.” Second, linking db/db signaling to protein synthesis pathways via the Stat3/Eif4e axis in iPSCs provides insights in understanding the incidence of cancer in leptin receptor dysfunctional states. Third, a decrease in the stem cell markers NOGGIN, NESTIN, and GFAP during directed differentiation of db/db iPSCs suggests a role for OB-R signaling in the regulation of neuronal development. Thus, db/db MEFs and iPSCs provide useful tools (depicted in the Graphical Abstract) to explore the relevance of OB-R in the early origins of obesity, diabetes, and metabolic abnormalities, and to find putative targets for harnessing therapeutics to prevent the development of these diseases.
Experimental Procedures
Mice and MEFs
All studies involving mice were approved by the institutional review board of the Joslin Diabetes Center and were in accordance with NIH guidelines. Embryonic day 14.5 wild-type control (Ctrl) (N = 4) and db/db MEFs (N = 4) were derived from breeding OB-Rdb/+ heterozygous mice (Jackson Laboratory). All fibroblasts were maintained up to a maximum passage ∼10 in Dulbecco modified Eagle's medium supplemented with GlutaMAX, 10% fetal bovine serum (FBS), and 1% nonessential amino acids.
Lentiviral-Mediated Reprogramming and iPSC Generation and Characterization
Generation of mouse iPSCs involved infection of primary MEFs with mouse STEMCCA lentivirus vector expressing the reprogramming factors Oct4, Sox2, Klf4, and cMyc. iPSC characterization involved teratoma formation, H&E staining, and immunostaining for the three lineage markers staining performed according to previous reports (Bhatt et al., 2015; Sommer et al., 2009; Teo et al., 2013). More information is available in the Supplemental material.
Gene Expression Analyses Using Quantitative RT-PCR and Western Immunoblotting
RNA was isolated from cells using RNeasy kit (QIAGEN) and cDNA was prepared from 1 μg of RNA using the RT-PCR kit (Life Technologies) according to the manufacturer's instructions and diluted to a final volume of 250 μL. In parallel experiments total cellular proteins were harvested using M-PER mammalian protein extraction reagent (Thermo Scientific) followed by western immunoblotting of proteins. More information about antibodies is included in Supplemental material.
Gene Correction in db/db iPSCs Using CRISPR-Cas9
The Cas9 and sgRNA plasmid pX459 were obtained from Addgene (plasmid no. 48139). Guide RNAs (20 bps) were chosen that lie in the intronic region between exons 18 and 19 of the OB-R gene using the CRISPR Design Tool (http://crispr.mit.edu/). The sequence of sgRNA cloned in vector was CACCGGATGTTTACATTTTGATGG (sgRNA-01), and a 200-nt DNA oligo complementing the region was used as repair template (seq CACTCTTTGAAGTCTCTCATGACCACTACAGATGAACCCAATCTACCAACTTCCCAACAGTCCATACAATATTAGAAGATGTTTACATTTTGATGGAGGGAAACAAACCTAAACTATGGTTTGAATGACTAAGAAATAACATTTGATGAGCTTATTAGAGAAGTGTATATTTTGTGGCCACAATGTAGGTTTGATGTAGT, purchased as ssODN Ultramer Oligo from IDT). db/db iPSCs cells were electroporated using 500 ng of vector and 50 pmol of ssODN template using Amaxa Nucleofector (cat. no. VAPH-1001, Nucleofector Program A-23). Cells were plated on a 15-cm dish and, after overnight culture, 50 ng/mL of puromycin selection was applied for the first 2 days of culture. Colonies started appearing within 5–7 days and individual colonies/clones were manually picked and transferred to 48-well plates.
Genotyping for 100 clones was performed by extracting genomic DNA (PureLink Genomic DNA Kit, Thermo Fischer Scientific) that was subjected to PCR (using F primer TCTCTGAATCCTTGCTCTTTGT, R primer CATGTGGTTGCTGGGATTTG) followed by Sanger sequencing using sequencing primer CCCTCTCCTAAGTGTGTCACTA. Data from sequencing traces were analyzed using SeqmanPro software (Lasergene SeqMan Pro version 8.1; DNASTAR). Clones with and without gene correction were compared for in vitro studies (isogeneic pair). In addition the clones were analyzed for potential off-target effects for top predicted off-target sites (NCBI-BLAST) using the Sanger sequencing method.
Flow Cytometry
iPSCs derived from Ctrl and db/db MEFs were trypsinized using 0.05% trypsin-EDTA. Single cells were stained for Mito-GFP dye with proper isotype controls using methods described previously (Gupta et al., 2015). For neuronal marker staining, cells were harvested and fixed in 4% paraformaldehyde for 15 min at room temperature. Cells were then spun and washed with cold fluorescence-activated cell sorting (FACS) buffer (5% FBS in PBS). Permeabilization and blocking was carried out on ice for 30 min in FACS buffer with 0.1% Triton X-100. Antibody staining was performed on ice for 30 min using chicken anti-NESTIN (1:100, NB100-1604; Novus Biologicals), rabbit anti-NOGGIN (1:50, ab-16054, Abcam) followed by incubation with secondary antibody (anti-chicken Alexa 488 and anti-rabbit Alexa 594; 1:1,000, Invitrogen) for 30 min on ice. Cells were washed and resuspended in 250 μL FACS buffer and filtered through a 30-μm filter before analysis by LSR II (BD Biosciences, Joslin Flow Cytometry Core). Gating was determined according to the secondary-only controls.
Metabolic Profiling
Bioenergetic profiles of MEFs and mouse iPSCs were generated using a Seahorse Bioflux Analyzer as reported earlier (Kleinridders et al., 2013). The brief method is described in the Supplemental material.
Chromatin Immunoprecipitation and Immunofluorescence
In brief, Ctrl and db/db iPSCs were fixed in 1% formaldehyde, and quenched in glycine. Cells were washed with PBS and lysed in cold lysis buffer with protease and phosphatase inhibitors (Sigma). Cells were then sonicated using 25% of power, 10-s on/off pulses for a total of 3 min. Immunoprecipitations were performed using anti-STAT3 antibody (Santa Cruz). Ctrl and db/db iPSCs were transferred onto poly-L-lysine pre-coated cover slips and incubated at 37°C for 24 h. Cells on cover slips were fixed with 2% formaldehyde and stained with anti-STAT3 antibody followed by anti-rabbit IgG Alexa 594 (Molecular Probes). Cover slips were mounted with Fluorescent Mounting Media (DAKO) and cells were analyzed under a Carl Zeiss LSM7 confocal laser scanning microscope.
Immunohistochemistry
Neuronal cells grown on glass slides were fixed in 4% paraformaldehyde solution (Wako) before staining with specific primary antibodies as described previously. More information is available in the Supplemental material.
Mass Spectrometry Analyses
We generated biological replicates for each of the db/db and Ctrl mouse cell cultures, both before (i.e., from MEFs; n = 2 + 2 replicates) and after reprogramming (i.e., from iPSCs; n = 4 + 4 replicates) as outlined in Figure S2. Subsequently, the eight iPSC and four MEF cell lines were multiplexed by isobaric labeling and subjected to independent liquid chromatography-mass spectrometry analysis as described in the Supplemental material.
RNA-Seq Analyses
We isolated total RNA from Ctrl (N = 4), db/db (N = 4), and db/Corr (N = 2) iPSCs in duplicates and sent it out to DNA Link for sequencing. RNA-seq data were analyzed in the Joslin Bioinformatic core facility. RNA-seq raw reads were downloaded from DNA Link, which were reverse stranded paired-end reads. There were 14,245 genes after filtering out the low expressing genes. We then normalized counts by weighted trimmed mean of M values. The normalization factors were between 0.93 and 1.08. Detailed methods for analyses are described in the Supplemental material.
Neuronal Differentiation
Fifty thousand Ctrl, db/db, and db/Corr iPSCs were plated on gelatin-coated six-well plates and differentiated up to 15 days in NDiff 227 medium (Clontech). Cells were harvested on day 15 for transcript and western blot analyses for neuronal markers.
Data and Code Availability
The accession number for the RNA-seq data reported in this paper is GSE155704 proteomics data reported in this paper via ProteomeXchange with identifier is PXD020959.
Author Contributions
M.K.G. conceived and designed the experimental study, and wrote the manuscript. H.V., S.S., T.N.R., V.W., J.S., H.B., M.V., T.T., S.K., M.S., R.M., J.H., and Y.B. performed the experiments and analyzed the data. H.R. supervised the study and edited the manuscript. R.N.K. conceived, designed, and supervised the study, provided funding and resources, and edited the manuscript. All authors read and agreed on manuscript.
Acknowledgments
We thank G. Mostoslavsky PhD (Boston University) for the kind gift of STEMCCA lentiviral plasmids and C.R. Kahn MD (Joslin Diabetes Center) for the use of the Seahorse instrument. We thank D.M. Smith PhD (Astra Zeneca) and B. Tyberg PhD (Astra Zeneca) for support and discussions, and G. Daley MD PhD (Children's Hospital, Boston) for discussions. The authors acknowledge Jonathan Dreyfuss and Hui Pan from Joslin Bioinformatics Core facility for RNA-seq analyses. The authors also acknowledge the iPSC Core Facility (DRC, NIH P30 DK036836), and C. Cahill for assistance with electron microscopy (Advanced Microscopy Facility DRC, NIH DK036836). M.K.G. is supported by a JDRF Advanced Postdoctoral Fellowship Award 3-APF-2017-393-A-N. S.S. is supported by NIH K12 HD000850, P30 DK40561, and NASPGHAN Young Investigator/Nestle Nutrition Award. J.S. is supported by a grant from SICORP of AMED with A∗STAR. R.N.K. acknowledges support from R01 DK67536 and R01 DK103215 and a grant from AstraZeneca. S.S is supported by NIH K12 HD000850. T.N.R. is supported by a grant from the Swiss National Science Foundation (320030_189090/1). H.B. and H.R are supported by The Bergen Research Foundation (Bergen Forskningsstiftelse); H.R. is supported by Novo Nordisk Fonden and Western Norway Regional Health Authority.
Published: October 29, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.10.001.
Supplemental Information
Table S6. Gene Stats in Ctrl iPSCs, db/db iPSCs and db/corr iPSCs
References
- Anisimov S.V., Christophersen N.S., Correia A.S., Hall V.J., Sandelin I., Li J.Y., Brundin P. Identification of molecules derived from human fibroblast feeder cells that support the proliferation of human embryonic stem cells. Cell Mol. Biol. Lett. 2011;16:79–88. doi: 10.2478/s11658-010-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S.H., Kulkarni R.N., Seifert M., Myers M.G., Jr. Roles for leptin receptor/STAT3-dependent and -independent signals in the regulation of glucose homeostasis. Cell Metab. 2005;1:169–178. doi: 10.1016/j.cmet.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bhatt S., Gupta M.K., Khamaisi M., Martinez R., Gritsenko M.A., Wagner B.K., Guye P., Busskamp V., Shirakawa J., Wu G. Preserved DNA damage checkpoint pathway protects against complications in long-standing type 1 diabetes. Cell Metab. 2015;22:239–252. doi: 10.1016/j.cmet.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M., Eckardt S., Scholer H.R., McLaughlin K.J. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott C.J., Johnson C.G., Yap C.C., Dwyer N.D., Litwa K.A., Winckler B. Nestin in immature embryonic neurons affects axon growth cone morphology and Semaphorin3a sensitivity. Mol. Biol. Cell. 2019;30:1214–1229. doi: 10.1091/mbc.E18-06-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen H., Charlat O., Tartaglia L.A., Woolf E.A., Weng X., Ellis S.J., Lakey N.D., Culpepper J., Moore K.J., Breitbart R.E. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- Chen N., Sugihara H., Kim J., Fu Z., Barak B., Sur M., Feng G., Han W. Direct modulation of GFAP-expressing glia in the arcuate nucleus bi-directionally regulates feeding. eLife. 2016;5 doi: 10.7554/eLife.18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K., Vaisse C., Lahlou N., Cabrol S., Pelloux V., Cassuto D., Gourmelen M., Dina C., Chambaz J., Lacorte J.M. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- Croizier S., Prevot V., Bouret S.G. Leptin controls parasympathetic wiring of the pancreas during embryonic life. Cell Rep. 2016;15:36–44. doi: 10.1016/j.celrep.2016.02.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.W., Camper S.A. Noggin regulates Bmp4 activity during pituitary induction. Dev. Biol. 2007;305:145–160. doi: 10.1016/j.ydbio.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hefnawy T., Ioffe S., Dym M. Expression of the leptin receptor during germ cell development in the mouse testis. Endocrinology. 2000;141:2624–2630. doi: 10.1210/endo.141.7.7542. [DOI] [PubMed] [Google Scholar]
- Ernst A., Sharma A.N., Elased K.M., Guest P.C., Rahmoune H., Bahn S. Diabetic db/db mice exhibit central nervous system and peripheral molecular alterations as seen in neurological disorders. Transl. Psychiatry. 2013;3:e263. doi: 10.1038/tp.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman D.E., Chen C., Punj V., Tsukamoto H., Machida K. Pluripotency factor-mediated expression of the leptin receptor (OB-R) links obesity to oncogenesis through tumor-initiating stem cells. Proc. Natl. Acad. Sci. U S A. 2012;109:829–834. doi: 10.1073/pnas.1114438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi N., Ziegler S., Wiestner A., Stoffel R., Heim M.H., Skoda R.C. Defective STAT signaling by the leptin receptor in diabetic mice. Proc. Natl. Acad. Sci. U S A. 1996;93:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gupta M.K., De Jesus D.F., Kahraman S., Valdez I.A., Shamsi F., Yi L., Swensen A.C., Tseng Y.H., Qian W.J., Kulkarni R.N. Insulin receptor-mediated signaling regulates pluripotency markers and lineage differentiation. Mol. Metab. 2018;18:153–163. doi: 10.1016/j.molmet.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.K., Teo A.K., Rao T.N., Bhatt S., Kleinridders A., Shirakawa J., Takatani T., Hu J., De Jesus D.F., Windmueller R. Excessive cellular proliferation negatively impacts reprogramming efficiency of human fibroblasts. Stem Cell Transl. Med. 2015;4:1101–1108. doi: 10.5966/sctm.2014-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.B., Mitchell T.D., Yan X., Simpson J.S., Redmann S.M., Jr Metabolic responses to leptin in obese db/db mice are strain dependent. Am J Physiol Regul Integr Comp Physiol. 2001 doi: 10.1152/ajpregu.2001.281.1.R115. [DOI] [PubMed] [Google Scholar]
- Hoggard N., Haggarty P., Thomas L., Lea R.G. Leptin expression in placental and fetal tissues: does leptin have a functional role? Biochem. Soc. Trans. 2001;29:57–63. doi: 10.1042/0300-5127:0290057. [DOI] [PubMed] [Google Scholar]
- Holmstrom M.H., Iglesias-Gutierrez E., Zierath J.R., Garcia-Roves P.M. Tissue-specific control of mitochondrial respiration in obesity-related insulin resistance and diabetes. Am. J. Physiol. Endocrinol. Metab. 2012;302:E731–E739. doi: 10.1152/ajpendo.00159.2011. [DOI] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinridders A., Lauritzen H.P., Ussar S., Christensen J.H., Mori M.A., Bross P., Kahn C.R. Leptin regulation of Hsp60 impacts hypothalamic insulin signaling. J. Clin. Invest. 2013;123:4667–4680. doi: 10.1172/JCI67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Forte T.M., Taniguchi S., Ishida B.Y., Oka K., Chan L. The db/db mouse, a model for diabetic dyslipidemia: molecular characterization and effects of Western diet feeding. Metabolism. 2000;49:22–31. doi: 10.1016/s0026-0495(00)90588-2. [DOI] [PubMed] [Google Scholar]
- Margetic S., Gazzola C., Pegg G.G., Hill R.A. Leptin: a review of its peripheral actions and interactions. Int. J. Obes. Relat. Metab. Disord. 2002;26:1407–1433. doi: 10.1038/sj.ijo.0802142. [DOI] [PubMed] [Google Scholar]
- Mohseni P., Sung H.K., Murphy A.J., Laliberte C.L., Pallari H.M., Henkelman M., Georgiou J., Xie G., Quaggin S.E., Thorner P.S. Nestin is not essential for development of the CNS but required for dispersion of acetylcholine receptor clusters at the area of neuromuscular junctions. J. Neurosci. 2011;31:11547–11552. doi: 10.1523/JNEUROSCI.4396-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Pellymounter M.A., Cullen M.J., Baker M.B., Hecht R., Winters D., Boone T., Collins F. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995 doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- Perez-Perez A., Maymo J., Gambino Y., Duenas J.L., Goberna R., Varone C., Sanchez-Margalet V. Leptin stimulates protein synthesis-activating translation machinery in human trophoblastic cells. Biol. Reprod. 2009;81:826–832. doi: 10.1095/biolreprod.109.076513. [DOI] [PubMed] [Google Scholar]
- Ramos-Lobo A.M., Teixeira P.D., Furigo I.C., Melo H.M., de M.L.E.S.N., De Felice F.G., Donato J., Jr. Long-term consequences of the absence of leptin signaling in early life. eLife. 2019;8 doi: 10.7554/eLife.40970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S., Bonnefond A., Manzoor J., Philippe J., Durand E., Arshad M., Sand O., Butt T.A., Falchi M., Arslan M. Novel LEPR mutations in obese Pakistani children identified by PCR-based enrichment and next generation sequencing. Obesity. 2014;22:1112–1117. doi: 10.1002/oby.20667. [DOI] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Gingras A.C. The mRNA 5' cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- Teo A.K., Windmueller R., Johansson B.B., Dirice E., Njolstad P.R., Tjora E., Raeder H., Kulkarni R.N. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J. Biol. Chem. 2013;288:5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse C., Halaas J.L., Horvath C.M., Darnell J.E., Jr., Stoffel M., Friedman J.M. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat. Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- Wang B., Chandrasekera P.C., Pippin J.J. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr. Diabetes Rev. 2014;10:131–145. doi: 10.2174/1573399810666140508121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S6. Gene Stats in Ctrl iPSCs, db/db iPSCs and db/corr iPSCs
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GSE155704 proteomics data reported in this paper via ProteomeXchange with identifier is PXD020959.