Figure 1.
Altered Metabolic Regulation in db/db MEFs and Reduced Pluripotency and Upregulated Protein Synthesis Pathway in db/db iPSCs
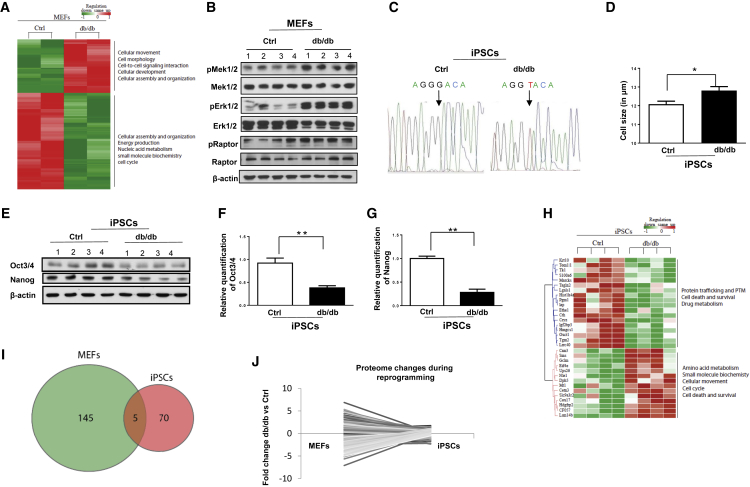
(A–G) (A) Heatmap representation of differentially expressed proteins between Ctrl and db/db MEFs. A selection of gene ontology terms is given for each protein cluster. Red indicates upregulated while green indicates downregulated proteins in db/db MEFs. (B) Western blots analysis showing upregulated MEK1/2, ERK1/2, and RAPTOR pathways in db/db MEFs. β-Actin was used as a loading control. (C) Genomic DNA sequencing showing G to T transition of base in db/db iPSCs. (D) Cell diameter of Ctrl and db/db iPSCs using Cello meter. (E) Western blot analysis of OCT4 and NANOG in four different clones from Ctrl and db/db iPSCs; quantification of changes in OCT4 (F) and NANOG (G) using ImageJ software. β-Actin was used as a loading control.
(H–J) (H) Heatmap representation of pathway analysis of differentially expressed proteins between Ctrl and db/db iPSCs. A selection of GO terms is given for each protein cluster. Red indicates upregulated while green indicates downregulated proteins in db/db iPSCs. (I) Venn diagram showing the numbers of proteins that were significantly differently expressed (p < 0.05) in db/db MEF-specific proteome and the db/db iPSC-specific proteome. Five proteins were significantly changed between db/db and Ctrl, both before and after reprogramming. (J) Pairing of protein expression values from each of the significant proteins in the db/db MEF-specific proteome with the db/db iPSC-specific proteome.