Figure 3.
Chromatin Immunoprecipitation Analysis for Stat3-Eif4e Binding and CRISPR Correction of the db/db Mutation in iPSCs
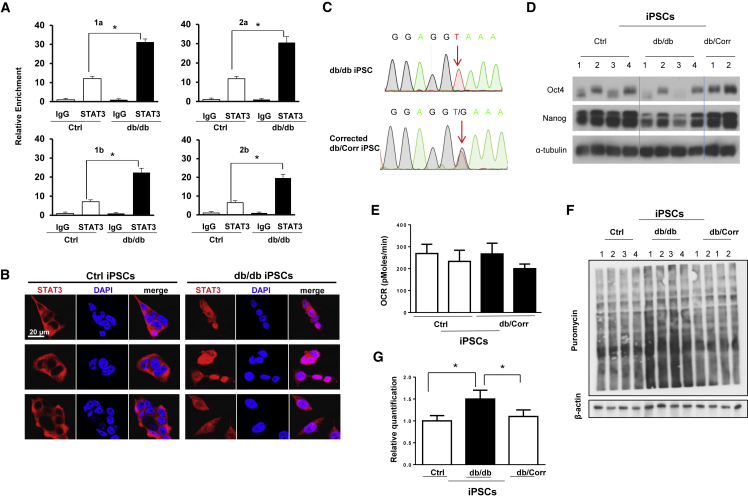
(A–E) (A) Chromatin immunoprecipitation analysis showing higher Stat3 binding (relative enrichment) on the two promoter regions “a” and “b” of Eif4e in db/db iPSCs using four different sets of primers 1a, 1b and 2a, 2b. (B) Live-confocal microscopy reveals the predominant nuclear localization of STAT3 in db/db iPSCs compared with cytosolic localization of STAT3 in Ctrl iPSCs. Scale bar, 20 μm. Each row indicates a single clone from Ctrl and db/db iPSCs. (C) db/db mutation (G to T) in the intronic region between exon 18 and exon 19, and DNA sequencing reveals the reversal of one copy to wild type in db/db iPSCs. Similar results were obtained in two additional clones. (D) Western blot analysis showing restoration of protein abundance of OCT4 and NANOG in CRISPR-corrected (db/Corr) iPSCs. α-Tubulin was used as a loading control. (E) Metabolic profiling showed similar basal respiration in db/db-corrected iPSCs compared with Ctrl iPSCs.
(F and G) (F) Anti-puromycin western blot analysis showing normalization of protein synthesis in CRISPR-corrected db/Corr iPSCs. β-Actin was used as a loading control. (G) Quantification of data from experiment in (F), n = 3 independent experiments; data are shown as mean ± SD and analyzed by Student's t test (∗p < 0.05, ∗∗p < 0.01).