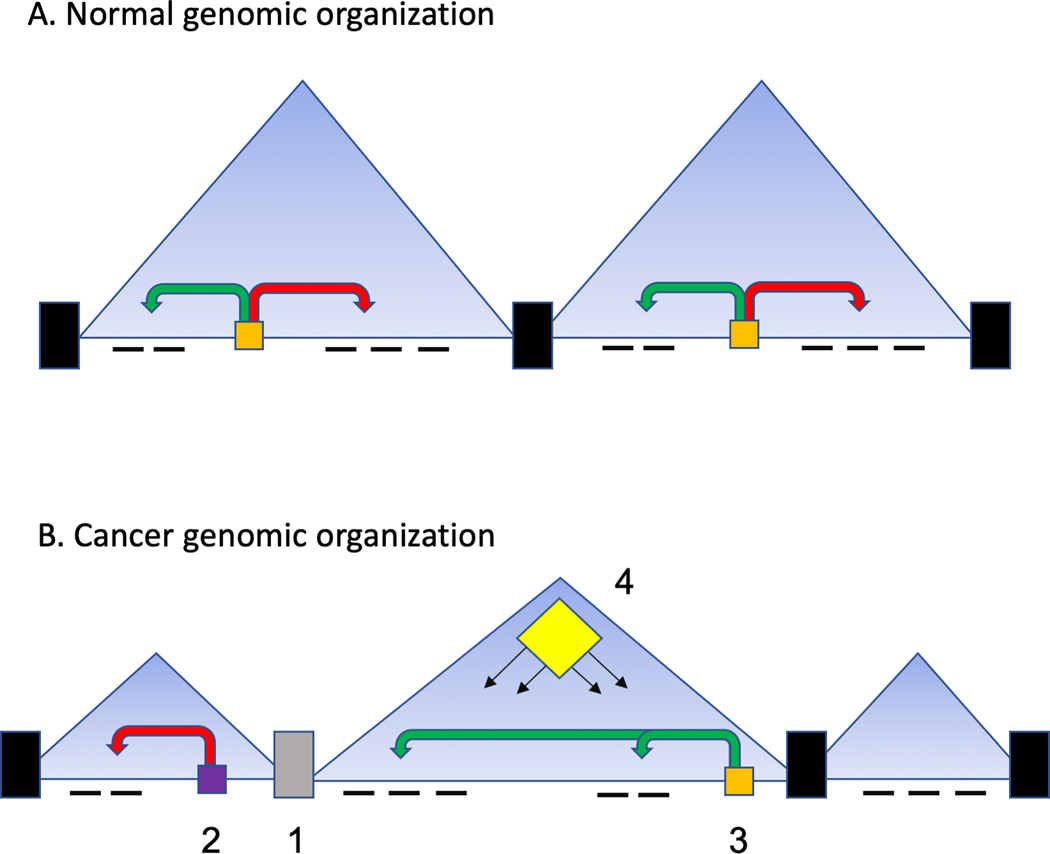
Figure 1: TAD organization in health and disease.
A. In normal cells TADs are organized such that boundaries arise by the action of multi-protein complexes centered on CTCF and cohesin complexes. These act to insulate the contents of the TAD which range in size from 100 kb to 1–2 mb. Within a TAD, enhancers (in orange) regulate target genes through further looping events and exert positive and negative impacts on gene expression. Often these gene expression patterns are highly coordinated and governed by multiple enhancer interactions.
B. In cancer cells there many examples of these process being corrupted. There is good evidence that boundary function is altered (1) with loss and gain of boundaries and changing the TAD structure. Generally, it appears the cancer genome gains more TADs of shorter length. This change of TAD structure can change enhancer function (2, 3) such that previously insulated genes are subsequently regulated by new enhancers. This is further distorted by structural variations impacting boundary function (1) and enhancer responses (2,3). The coordinated gene regulation within a TAD also allows for emergent changes in epigenetic regulation (4). That is, mutations in epigenetic regulators such as EZH2 can modestly change the regulation of individual genes in a TAD, but the collective effect on all genes is pronounced if all the genes are on a pathway for example.