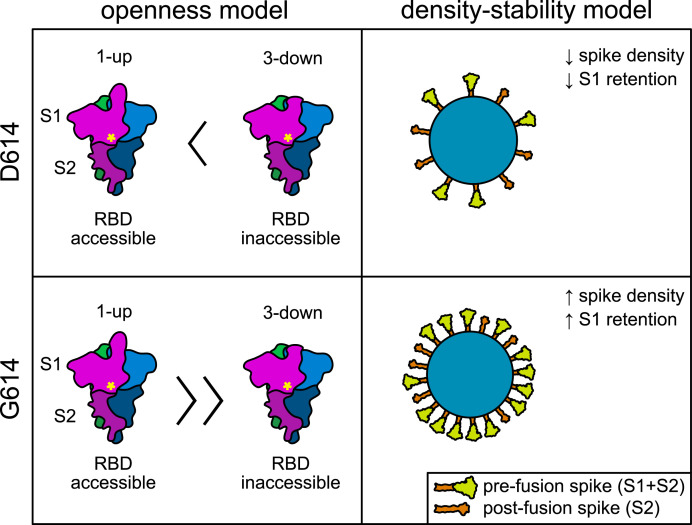
Fig. 2.
Schematic comparison of two independent models explaining increased infectivity by SARS-CoV-2 isolates possessing D614G mutant spike. On the left, the “openness” model explains increased infectivity by an increased propensity of S(G614) spike to assume the 1-up conformation thought to be necessary for ACE2 interaction with the RBD. Magenta, blue, and green colors represent individual S protein monomers. Yellow asterisk indicates position of residue 614 near the S1/S2 interface. On the right, the “density-stability” model explains increased infectivity by increased stability of S(G614) spike trimers, facilitating greater incorporation into virions and less shedding of the S1 subunit. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)