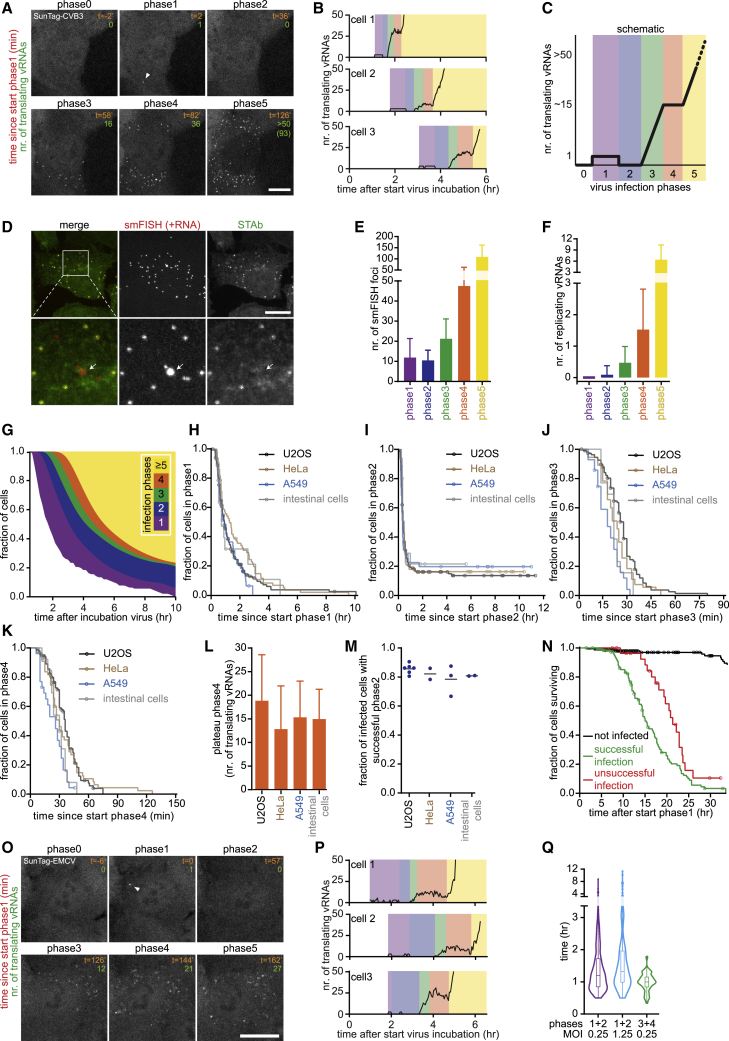
Figure 2.
Single-Cell Dynamics and Heterogeneity of Virus Replication
(A and B) Representative images (A) and example quantifications (B) of time-lapse movies of STAb cells infected with SunTag-CVB3. Example images from the same time-lapse movie are also used in Figure 1G and Video S1.
(C) Cartoon of infection phases in single cells.
(D) Representative images of vRNA smFISH and STAb staining.
(E and F) Combined analysis of live-cell imaging to determine infection phase and smFISH in the same cells to quantify smFISH foci (E) and replicating vRNAs (F) in STAb cells infected with SunTag-CVB3.
(G) Fraction of cells in each infection phase over time. Uninfected cells are excluded from quantification.
(H–K) Kaplan-Meier graphs showing durations of infection phases.
(L) Mean number of translating vRNAs during phase 4.
(M) Fraction of infected cells with successful phase 2, based on the plateau in the Kaplan-Meier curve of the duration of phase 2. Every dot indicates a repeat, and lines indicate mean.
(N) Kaplan-Meier survival curve.
(O and P) Representative images (O) and example quantifications (P) of representative time-lapse movie of STAb cells infected with SunTag-EMCV.
(Q) Violin plot of the combined timing of phases 1 and 2 or phases 3 and 4.
Colors (B, C, and P) illustrate infection phases. Data points during phases 0, 1, and 2 (B and P) are increased 3-fold to aid visualization of data that are very close to the x axes. Arrowheads (A and E) indicate the first translating vRNA; an arrow (D) indicates a replicating vRNA. Error bars (E, F, and L) indicate SD; circles (H–K and N) indicate the last analyzable time point for individual cells. Scale bars, 15 μm. See also Videos S1, S2, and S3 and Figures S2 and S3. The number of experimental repeats and cells analyzed per experiment are listed in Table S1.