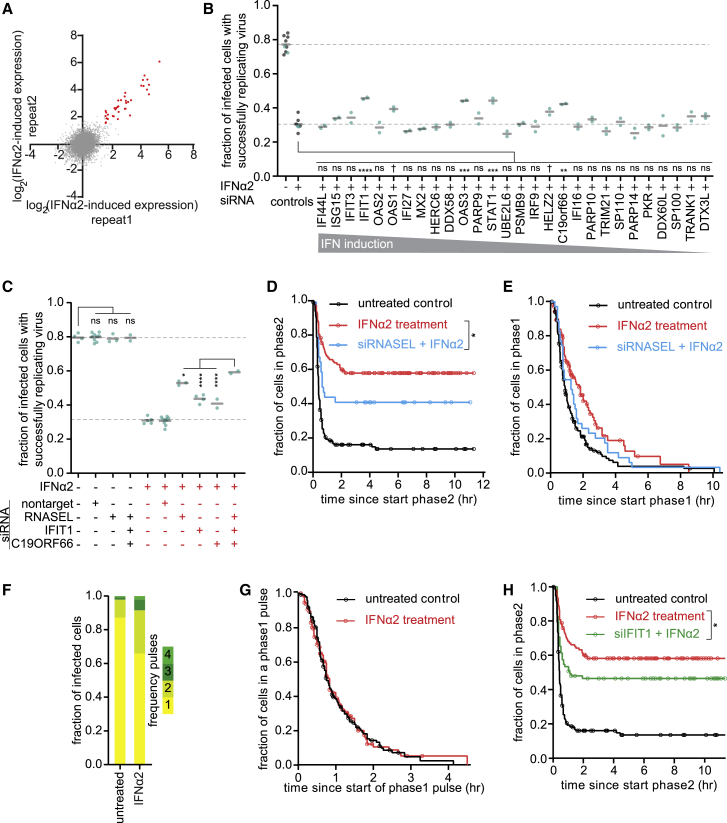
Figure 6.
IFN Acts through Multiple Parallel Mechanisms to Block Replication of the Incoming vRNA
(A) Scatterplot of gene expression changes after IFNα2 treatment. Every dot indicates a single gene, and red dots indicate genes with at least 3-fold increased expression upon IFNα2 treatment in both repeats.
(B and C) Fraction of infected cells with successfully replicating viruses after transfection with the indicated siRNAs and/or treatment with IFNα2. Every dot represents an independent experiment, and black lines indicate the means. Grey control data are based on experiments without siRNA transfections; blue control data are based on non-targeting siRNAs. Dashed lines represent the means of controls.
(D and E) Kaplan-Meier graphs showing the duration of infection phase 2 (D) or phase 1 (E). Data in black and red are replotted from Figures 5B and 5C for comparison.
(F) Frequency of pulses. Control data are replotted from Figure 3C.
(G) Kaplan-Meier graphs showing the duration of phase 1 pulses. Data in black are replotted from Figure S3D for comparison.
(H) Kaplan Meier graphs showing the duration of infection phase 2. Data in black and red are replotted from Figure 5C for comparison.
Circles (D, E, G, and I) indicate the last analyzable time point for individual cells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01, p < 0.001, ∗∗∗∗p < 0.0001; Dunnett’s multiple comparisons test (B and C) or Gehan-Breslow-Wilcoxon test (D and H). † indicates non-significant genes that were included in the follow-up analysis. See also Figure S6. The number of experimental repeats and cells analyzed per experiment are listed in Table S1.