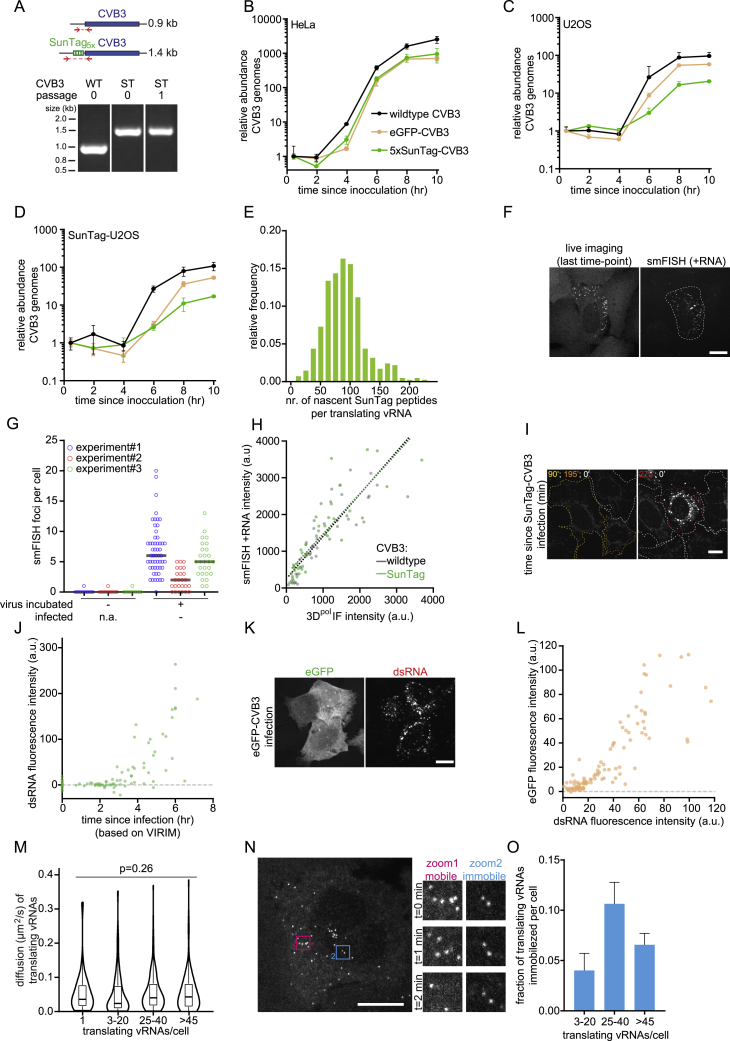
Figure S1.
A Single-Molecule Assay to Visualize and Analyze Translation and Replication of Individual CVB3 vRNAs, Related to Figure 1
(A) Agarose gel analysis of the SunTag insert in the wild-type (WT) or SunTag (ST) CVB3 genome after RNA isolation, cDNA synthesis and PCR amplification of the indicated region. (B-D) Virus growth curves of the indicated viruses in indicated cell lines. Similar MOIs were used in all experiments. (E) Number of nascent SunTag peptides per translating vRNA focus based on the vRNA GFP fluorescence intensities compared to the GFP fluorescence intensity of 24xSunTag arrays expressed in STAb cells. (F) Representative images of STAb cell infected with SunTag-CVB3, during live-cell imaging (left) or after fixation and smFISH against +CVB3. (G) The number of smFISH +CVB3 ‘background’ foci in cells not exposed to virus (left) or cells that were uninfected after incubation with SunTag-CVB3 (right). For each repeat, the mean number of smFISH foci in uninfected cells was used to correct the number of smFISH foci in infected cells in Figures 1E, 1I, and 2E. (H) Combined analysis of viral protein synthesis (based on 3Dpol protein immunofluorescence) and viral load (based on fluorescence intensity of +CVB3 smFISH) in the STAb cells infected with indicated virus. Dashed lines indicate linear fits. (I, J) Representative images (I) and quantification (J) of combined analysis of live-cell imaging and dsRNA immunofluorescence of the same STAb cells infected with SunTag-CVB3. Color of outline (I) indicates the time between first detection of a translating vRNA and fixation. Cells in which no translating vRNAs were observed are indicated by a white outline and were used to correct for background fluorescence. (K, L) Representative images (K) and quantification (L) of combined analysis of GFP fluorescence and dsRNA immunofluorescence in the same U2OS cells infected with eGFP-CVB3. (M) Violin and boxplots of diffusion kinetics of translating vRNAs in cells that contain the indicated number of translating vRNAs. (N) Images of representative time-lapse movie of a STAb U2OS cell infected with SunTag-CVB3. Zooms indicate areas with mobile (pink) or immobilized (blue) translating vRNAs. (O) Bar graph of the fraction of immobilized translating vRNAs per cell. Every dot (G, H, J, L) indicates a single cell. Statistics is based on Kruskal-Wallis test. Error bars indicate SEM. Scale bars, 15 μm. The number of experimental repeats and cells analyzed per experiment are listed in Table S1.