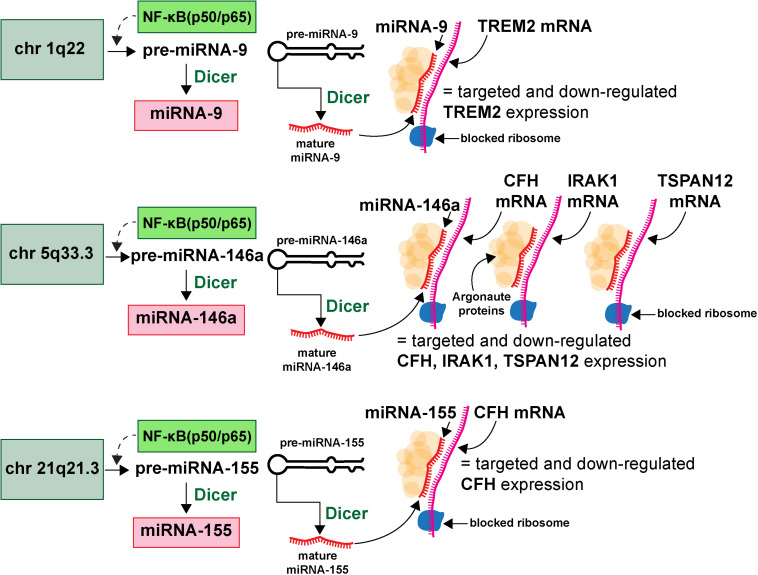
FIGURE 2.
Highly simplified schematic of an AD-relevant microRNA-messenger RNA (miRNA-mRNA) regulatory network involving 3 different miRNAs and 4 different mRNAs; this drawing graphically illustrates the molecular-genetic mechanism of microRNA (miRNA) generation and targeted miRNA-mRNA interaction; selective pathogenic families of miRNAs (for example miRNA-9, miRNA-146a and miRNA-155), transcribed from genes located on 3 different chromosomes (chr 1q22, chr 5q33.3, chr 21q21.3) generate precursor miRNAs (pre-miRNAs) which are subsequently processed into neurologically active mature miRNAs (miRNA-9, miRNA-146a and miRNA-155 shown as an example); many more chromosomes, miRNAs and mRNAs and miRNA-mRNA signaling networks are probably involved; many AD-relevant miRNA encoding genes are under transcriptional control by the pro-inflammatory transcription factor NF-kB (p50/p65); mature miRNAs subsequently find their target mRNAs (TREM2, CFH, IRAK-1, and TSPAN12 shown) and the miRNA-mRNA double-stranded RNA complex is blocked at the entrance to the ribosome (blue spherical complex on mRNA stand) and the miRNA-mRNA complex is degraded; the major mode of miRNA action in the mammalian brain is pathologically up-regulated miRNAs driving the down-regulation of AD-relevant genes (see text); single miRNAs can target multiple mRNAs and multiple miRNAs can target a single mRNA (see also Figure 3); miRNAs have established roles in recognizing multiple mRNA sequences (genetic pleiotropy), combinatorial and cooperativity in gene regulation, template accessibility (mediated by various RNA binding proteins; in this diagram orange spheres at the miRNA-mRNA interface called “Argonaute proteins”) and post-transcriptional regulation of the transcriptome (Hobert, 2008; Jaber et al., 2019; Eisen et al., 2020; Lukiw, 2020a,b). Combined with other metrics, the precise quantitation of miRNA abundance, speciation and complexity in various AD biofluids has strong potential for increasing the accuracy of AD diagnostics; recent preliminary in vitro studies further indicate that anti-miRNA (antimiR, antagomir, AM)-based therapies may be effective in quenching the excessive miRNA-mediated downregulation of critical mRNA-driven gene expression in AD (Zhao et al., 2016a,b; Jaber et al., 2019; Fan et al., 2020; Ghaffari et al., 2020).