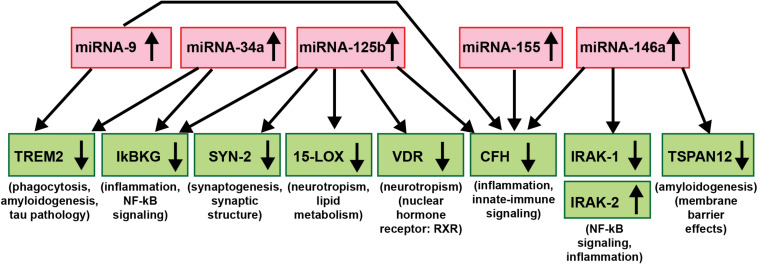
FIGURE 3.
An example of a highly interactive miRNA-mRNA regulatory network involving 5 miRNAs and 9 mRNAs; there is a significant contribution of up-regulated miRNA signaling (red boxes) to specific aspects of the down-regulated mRNA-mediated expression of key AD-relevant genes (green boxes) and AD neuropathology (Chandrasekaran and Bonchev, 2016; Clement et al., 2016; Hill and Lukiw, 2016; Jaber et al., 2017, 2019; Lukiw, 2020a,b). Just 5 significantly up-regulated miRNAs – miRNA-9, miRNA-34a, miRNA-125b, miRNA-146a, and miRNA-155 (all reported to be up-regulated in AD and/or TgAD models) – can account for the down-regulation of 8 mRNA targets critically involved in multiple aspects of AD neuropathology (IRAK-2 is up-regulated due to a compensatory mechanism as described in Cui et al., 2010). Briefly, these miRNAs down-regulate miRNA-directed mRNA target degradation involved in phagocytosis deficits, amyloidogenesis and tau pathology (TREM2, TSPAN12), inflammation, NF-kB- and innate-immune signaling (IkBKG, CFH, IRAK1; with a compensatory increase in IRAK-2), neurotropism (15-LOX, VDR), synaptic maintenance and synaptogenesis (SYN-2), all of which are distinguishing pathological features characteristic of AD neuropathology. Using DNA and RNA sequencing, microfluidic-based GeneChip microarray analysis and advanced LED-Northern dot blot analysis it has been recently reported that: (i) miRNA-9, miRNA-34a, miRNA-125b, miRNA-146a and miRNA-155 are easily detected in the human brain neocortex and retina; (ii) all have NF-kB-recognition features in their immediate upstream promoters; (iii) these same miRNAs are induced by increases in NF-kB due to reactive-oxygen species (ROS) induced stress; and (iv) these 5 miRNAs form a pro-inflammatory gene family up-regulated in AD brain neocortex and hippocampal CA1 (Colangelo et al., 2002; Cogswell et al., 2008; Zhao et al., 2015; Fan et al., 2020; Lukiw, 2020a,b). This diagram is based on studies from our laboratories in which each AD and age-and gender-matched control sample (N∼135) were interrogated for 2,650 human miRNAs and 27,000 human mRNAs using RNA sequencing, microarray analysis and/or advanced LED-Northern dot blotting technologies in a single experiment and miRNA-mRNA linkage analysis and bioinformatics were subsequently analyzed (Jaber et al., 2019; Fan et al., 2020; Lukiw and Pogue, 2020; manuscript in preparation).