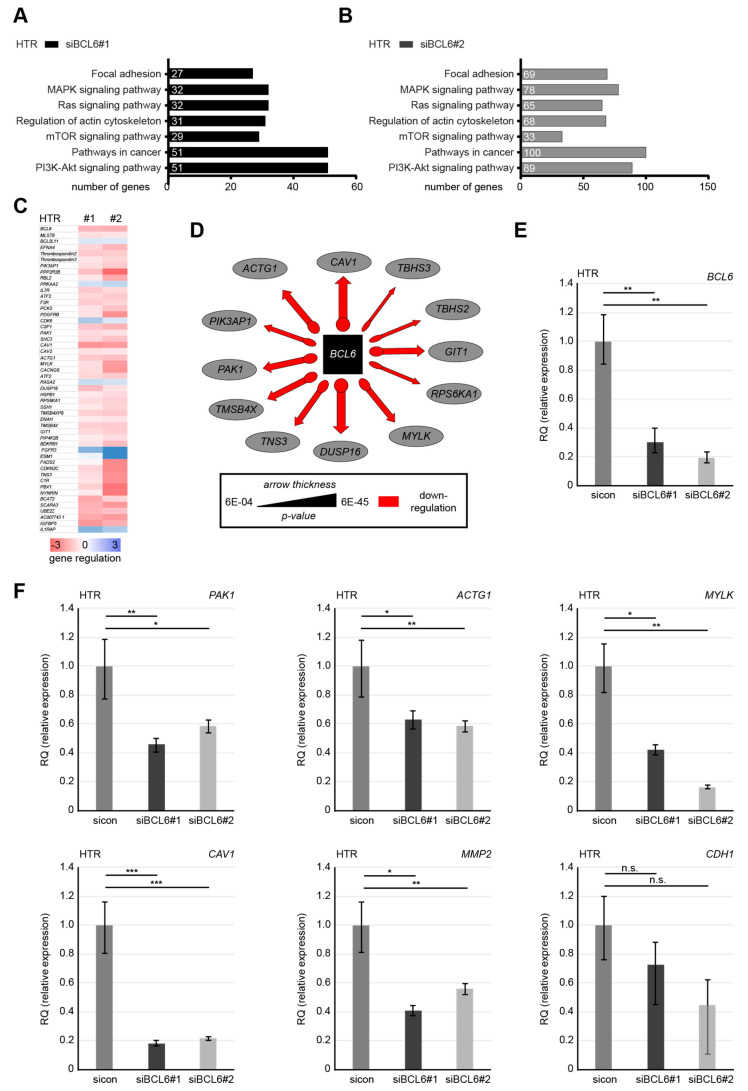
Figure 7.
Reduction of BCL6 reduces the levels of multiple genes related to migratory machinery. (A and B) HTR cells were treated with sicon, siBCL6#1, or siBCL6#2 for 48 h and the RNA from three independent experiments was extracted for transcriptome analysis. The effects of siBCL6#1 (A) or siBCL6#2 (B) on different pathways were determined by gene ontology enrichment analysis using KEGG pathway enrichment analysis. (C) A heatmap of the most differently expressed genes. Gene expression was analyzed using the DESeq2 R package. Genes with a p-value < 0.01 and a fold change greater than 3 (blue color code) and below 3 (red color code), respectively, are included. (D) The most significant genes affected by the depletion of BCL6 in the context of migration are depicted. (E) BCL6 gene analysis as the transfection efficiency control. (F) The gene levels of p21 RAC1 activated kinase 1 (PAK1), actin gamma (ACTG1), myosin light chain kinase (MYLK), caveolin 1 (CAV1), and matrix metalloproteinase-2 (MMP2) were measured. The mRNA data are based on three independent experiments and presented as the RQ, with the minimum and maximum range. RQ: relative quantification of gene expression. * p < 0.05, ** p < 0.01, and *** p < 0.001.